Search Count: 27
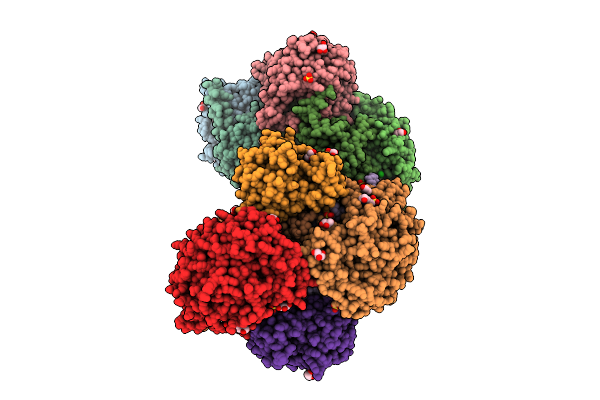 |
Dimeric State Of The F420-Reducing Hydrogenase From Methanothermococcus Thermolithotrophicus In Crystalline Form 1
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: NFU, NA, SO4, GOL, FE, SF4, FAD, CL, EPE |
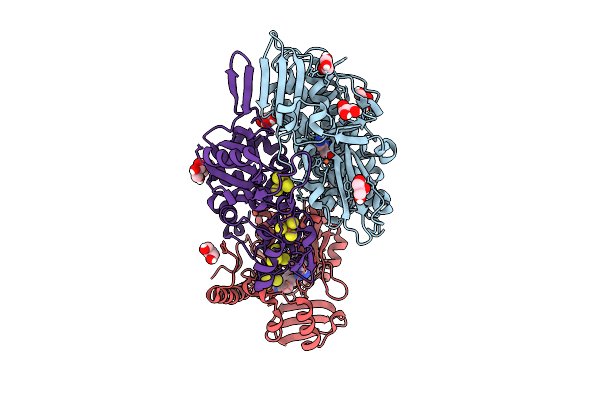 |
Dimeric State Of The F420-Reducing Hydrogenase From Methanothermococcus Thermolithotrophicus In Crystalline Form 2
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: NFU, GOL, FE, SF4, FAD |
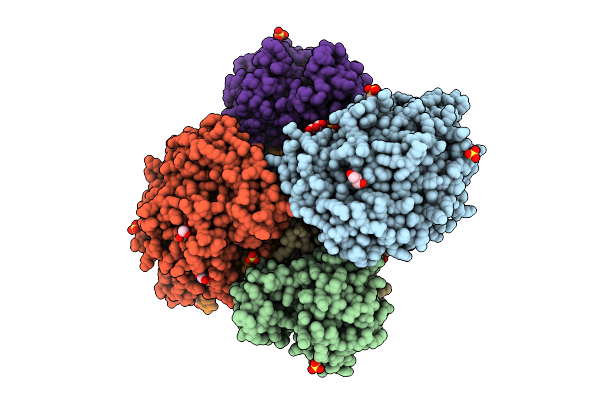 |
Dimeric State Of The F420-Reducing Hydrogenase From Methanothermococcus Thermolithotrophicus In Crystalline Form 3
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: GOL, MES, NFU, FE, SO4, EDO, FAD, SF4, PE4, FES |
 |
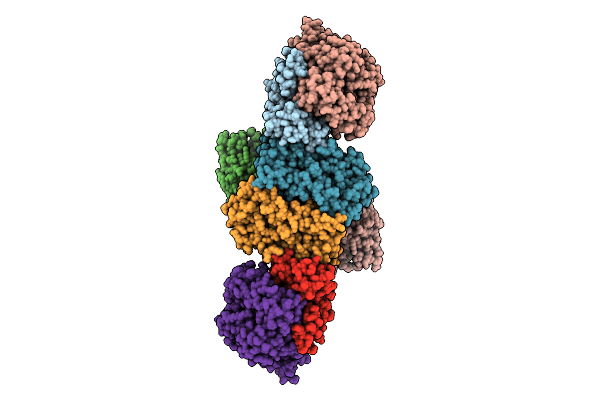
Cubic State Of The F420-Reducing Hydrogenase From Methanothermococcus Thermolithotrophicus
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: SO4, NFU, FAD, GOL, SF4 |
 |
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, FAD, NFU |
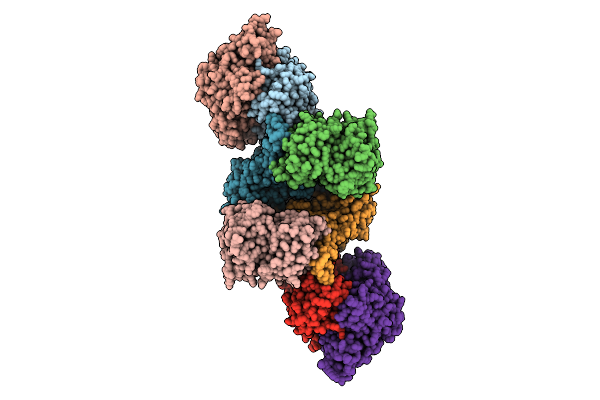 |
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, FAD, NAP, NFU, MG |
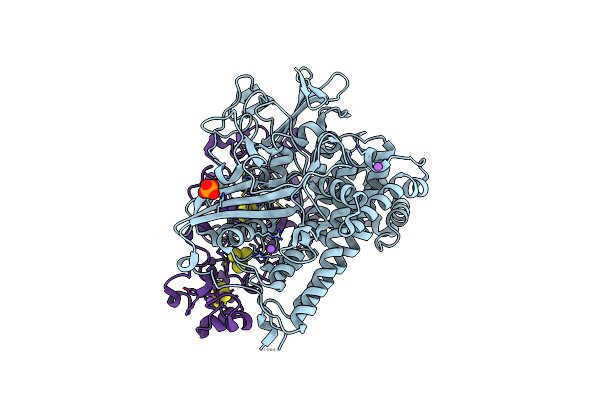 |
Crystal Structure Of The C19G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The H2-Reduced State At 1.6 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-09-18 Classification: OXIDOREDUCTASE Ligands: NFU, MG, NA, PO4, SF4, F3S, CL, ER2 |
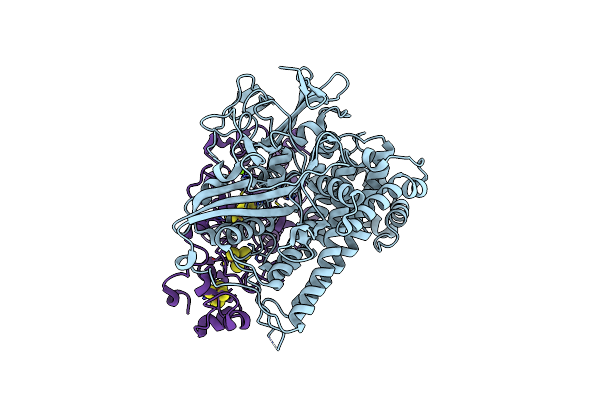 |
Crystal Structure Of The C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The H2-Reduced State At 1.65 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-09-18 Classification: OXIDOREDUCTASE Ligands: NFU, MG, CL, SF4, F3S, 35L, F4S |
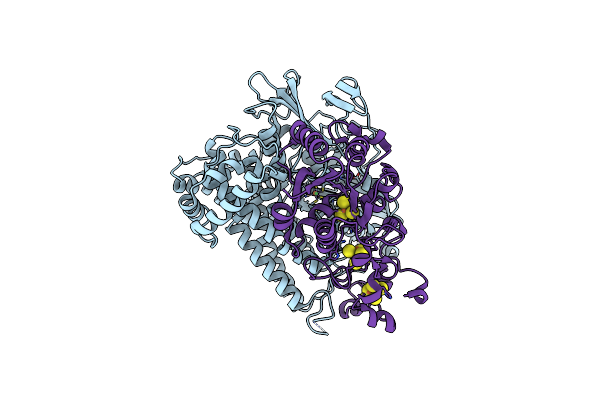 |
Crystal Structure Of The C19G/C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The H2-Reduced State At 1.92 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-11-01 Classification: OXIDOREDUCTASE Ligands: NFU, MG, SF4, F3S |
 |
Neutron Structure Of [Nife]-Hydrogenase From D. Vulgaris Miyazaki F In Its Oxidized State
Organism: Desulfovibrio vulgaris str. 'miyazaki f'
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.04 Å, 2.20 Å Release Date: 2023-09-13 Classification: OXIDOREDUCTASE Ligands: NFU, MPD, OH, MG, CL, SF4, F3S |
 |
The Carbon Monoxide Complex Of [Nife]-Hydrogenase (Hyb-Type) From Citrobacter Sp. S-77
Organism: Citrobacter sp. s-77
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2022-02-09 Classification: OXIDOREDUCTASE Ligands: MG, NFU, CMO, GOL, SF4, F3S |
 |
Crystal Structure Of The O2-Tolerant Mbh-P242C From Ralstonia Eutropha In Its Reduced State
Organism: Cupriavidus necator (strain atcc 17699 / h16 / dsm 428 / stanier 337), Cupriavidus necator (strain atcc 17699 / dsm 428 / kctc 22496 / ncimb 10442 / h16 / stanier 337)
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2021-07-07 Classification: ELECTRON TRANSPORT Ligands: NFU, MG, SF4, F4S, CL |
 |
The F420-Reducing [Nife] Hydrogenase Complex From Methanosarcina Barkeri At The Nia-S State
Organism: Methanosarcina barkeri ms
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-10-23 Classification: OXIDOREDUCTASE Ligands: SF4, FES, 144, BU3, MPD, NFU, FE, MG, FAD |
 |
Xenon Derivatization Of The F420-Reducing [Nife] Hydrogenase Complex From Methanosarcina Barkeri
Organism: Methanosarcina barkeri ms
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2019-10-23 Classification: OXIDOREDUCTASE Ligands: SF4, 144, FES, XE, BU3, NFU, FE, MG, FAD |
 |
Crystal Structure Of Aerobically Purified And Anaerobically Crystallized D. Vulgaris Miyazaki F [Nife]-Hydrogenase
Organism: Desulfovibrio vulgaris (strain miyazaki f / dsm 19637)
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2018-08-08 Classification: OXIDOREDUCTASE Ligands: SF4, F3S, MPD, MRD, TRS, MG, NFU |
 |
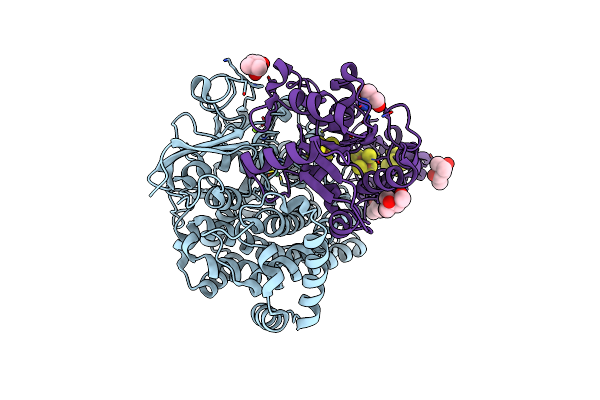
[Nife]-Hydrogenase (Hyb-Type) From Citrobacter Sp. S-77 In An H2-Reduced Condition
Organism: Citrobacter sp. s-77
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2018-06-27 Classification: OXIDOREDUCTASE Ligands: MG, NFU, GOL, SF4, F3S |
 |
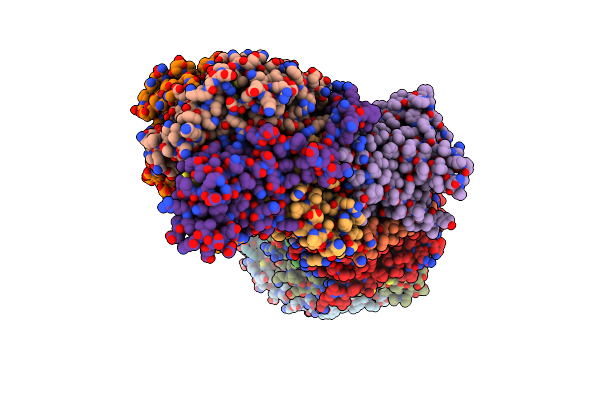
Crystal Structure Of Anaerobically Purified And Anaerobically Crystallized D. Vulgaris Miyazaki F [Nife]-Hydrogenase
Organism: Desulfovibrio vulgaris (strain miyazaki f / dsm 19637)
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2018-06-06 Classification: OXIDOREDUCTASE Ligands: SF4, F3S, MPD, TRS, NFU, MG |
 |
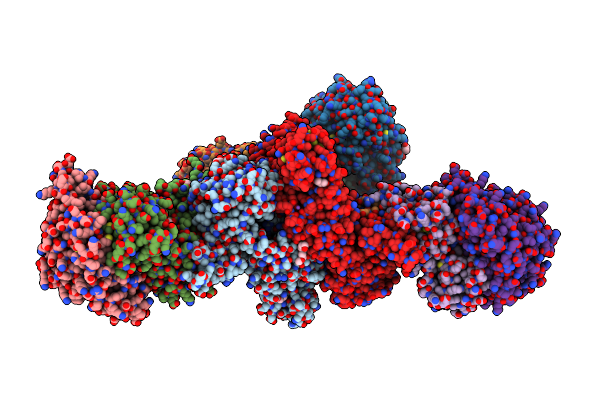
Organism: Pyrococcus furiosus com1, Pyrococcus furiosus (strain atcc 43587 / dsm 3638 / jcm 8422 / vc1)
Method: ELECTRON MICROSCOPY Resolution:3.70 Å Release Date: 2018-05-23 Classification: MEMBRANE PROTEIN Ligands: SF4, NFU |
 |
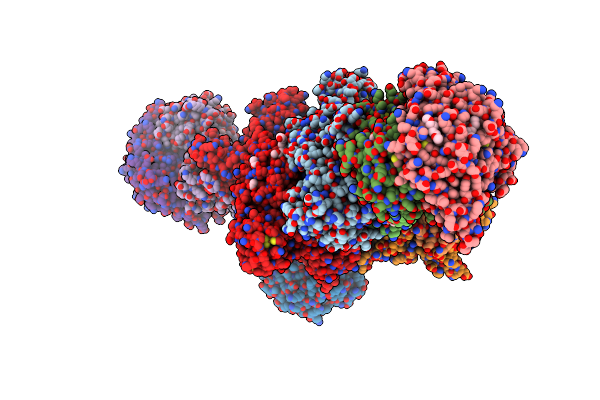
Heterodisulfide Reductase / [Nife]-Hydrogenase Complex From Methanothermococcus Thermolithotrophicus At 2.3 A Resolution
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-08-30 Classification: OXIDOREDUCTASE Ligands: FAD, NA, ACT, PE3, SF4, GOL, 9S8, FES, TRS, CA, MG, NFU |
 |
Heterodisulfide Reductase / [Nife]-Hydrogenase Complex From Methanothermococcus Thermolithotrophicus Soaked With Heterodisulfide For 3.5 Minutes
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-08-30 Classification: OXIDOREDUCTASE Ligands: SF4, GOL, PE3, FAD, 9S8, COM, FES, TRS, NFU, FE, ACT, TP7 |

