Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Structure Of Human Udp-Galactose 4-Epimerase In Complex With Covalent Compound Wbc10
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: NAI, A1IU6 |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: FAD, NAI |
 |
Crystal Structure Of Ni2+ Dependent Glycerol-1-Phosphate Dehydrogenase Aram From Bacillus Subtilis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: 13P, NAI, NI |
 |
Trypanosoma Cruzi Beta-3-Hbdh R19T/K20S/C64Y Mutant In Complex With Nadh And Malonate (C2 Space Group)
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: MLI, NAI, EDO |
 |
Trypanosoma Cruzi R19T/K20S/C64Y Mutant Beta-3-Hbdh Structure In Complex With Nadh And Malonate (P1 Space Group)
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: MLI, EDO, NAI |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: ACY, GOL, ZN, NAI |
 |
Horse Liver Alcohol Dehydrogenase F93W In Complex With Nadh And N-Cylcohexyl Formamide
Organism: Equus caballus
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, CXF |
 |
Horse Liver Alcohol Dehydrogenase V203L In Complex With Nadh And N-Cylcohexyl Formamide
Organism: Equus caballus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, CXF |
 |
Horse Liver Alcohol Dehydrogenase V203A In Complex With Nadh And N-Cylcohexyl Formamide
Organism: Equus caballus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, CXF |
 |
Electron-Bifurcating Tungstopyranopterin-Containing Aldehyde Oxidoreductase With Nadh
Organism: Acetomicrobium mobile
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: OXIDOREDUCTASE Ligands: SF4, FES, ZN, FMN, NAI, PTE, MG |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, CA, FES, FAD, NAI |
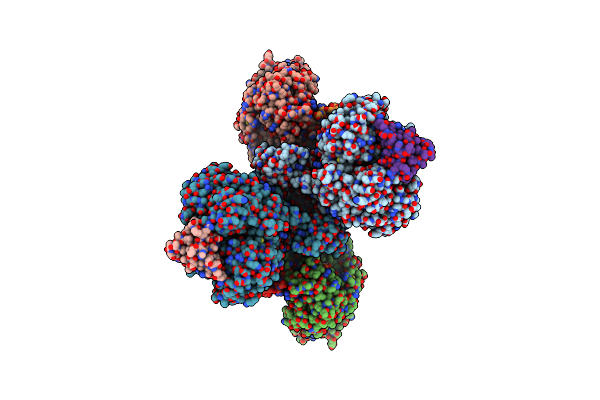 |
Cryo-Em Structure Of Reduced Form Of Formate Dehydrogenase From Rhodobacter Aestuarii (Rafdh) With Nadh
Organism: Rhodobacter aestuarii
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: OXIDOREDUCTASE Ligands: MGD, 6MO, FES, SF4, NAI, FMN |
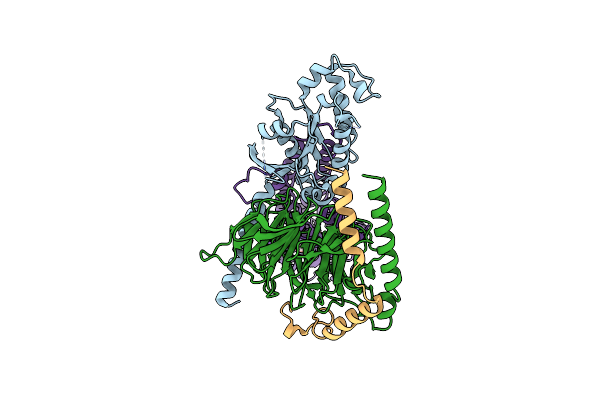 |
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: NAI |
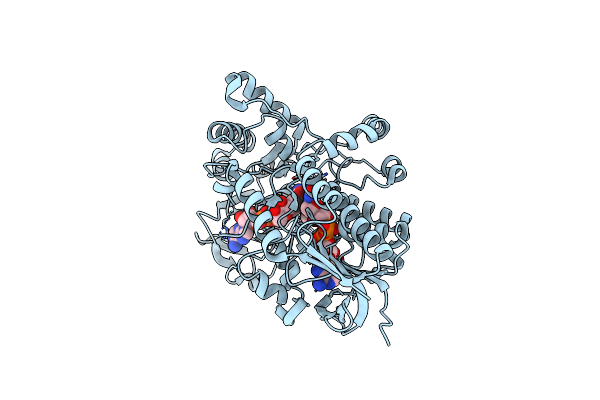 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: NAI, FAD |
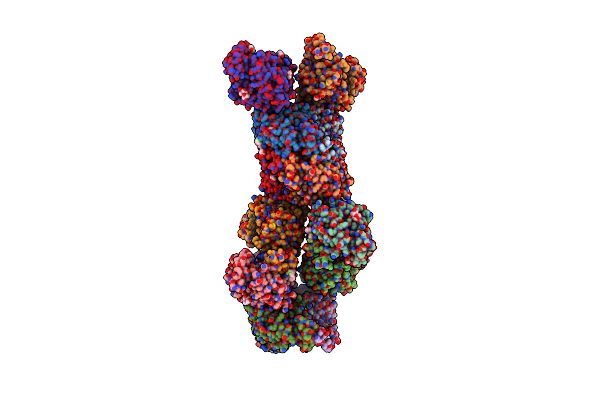 |
Crystal Structure Of Human L-Lactate Dehydrogenase B Protein In Complex With Nadh, Oxamate And Fluoxetine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: SFX, OXM, NAI, BTB, SO4, GOL, PEG |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: FAD, NAI |
 |
Crystal Structure Of Short Chain Dehydrogenase Reductase 9 (Short Chain Dehydrogenase Reductase 9C4) In Complex With Nadh
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: NAI |
 |
Tad From Carmabin Biosynthetic Pathway In Complex With Nadh - Crystal Form 1
Organism: Moorena producens 3l
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: NAI |
 |
Organism: Streptomyces albogriseolus
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2024-12-25 Classification: FLAVOPROTEIN Ligands: FAD, NAI |
 |
Structure Of The Enoyl-Acp Reductase Fabv From Pseudomonas Aeruginosa With Nadh Cofactor
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: NAI |