Search Count: 21
 |
Organism: Myxococcus xanthus dk 1622
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-06-04 Classification: SIGNALING PROTEIN Ligands: EDO |
 |
Organism: Myxococcus xanthus dk 1622
Method: X-RAY DIFFRACTION Release Date: 2025-03-26 Classification: REPLICATION Ligands: MG, ATP |
 |
Cargo-Loaded Myxococcus Xanthus Enca Encapsulin Engineered Pore Mutant With T=4 Icosahedral Symmetry
Organism: Myxococcus xanthus dk 1622
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: VIRUS LIKE PARTICLE |
 |
Myxococcus Xanthus Enca Encapsulin Engineered Pore Mutant With T=1 Icosahedral Symmetry
Organism: Myxococcus xanthus dk 1622
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: VIRUS LIKE PARTICLE |
 |
Cargo-Loaded Myxococcus Xanthus Enca Encapsulin Engineered Pore Mutant With T=3 Icosahedral Symmetry
Organism: Myxococcus xanthus dk 1622
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Myxococcus xanthus dk 1622
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: VIRUS LIKE PARTICLE |
 |
Myxococcus Xanthus Enca Protein Shell With Compartmentalized Snap-Tag Cargo Protein
Organism: Myxococcus xanthus dk 1622, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Myxococcus xanthus dk 1622
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CELL ADHESION |
 |
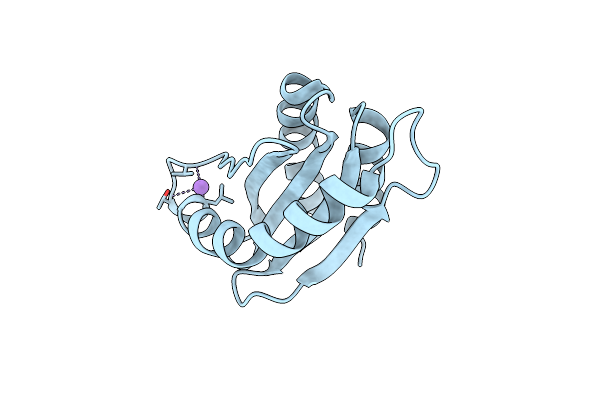
Organism: Myxococcus xanthus dk 1622
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-01-27 Classification: CYTOSOLIC PROTEIN |
 |
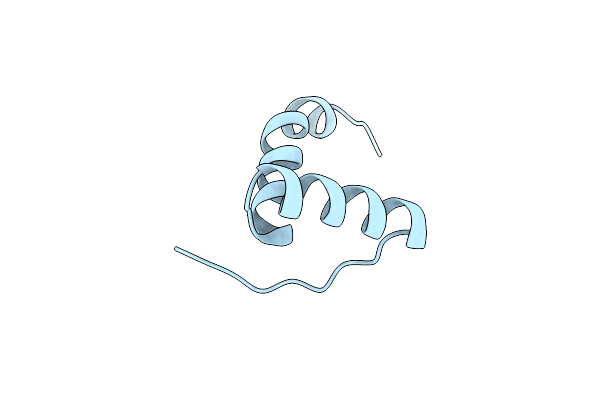
Organism: Myxococcus xanthus dk 1622
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2021-01-27 Classification: CYTOSOLIC PROTEIN |
 |
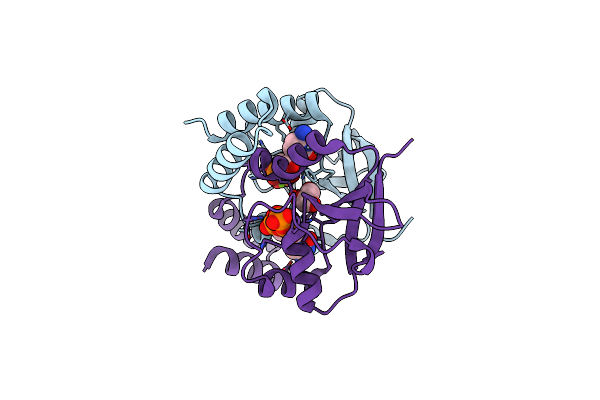
Organism: Myxococcus xanthus dk 1622
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2020-04-22 Classification: DNA BINDING PROTEIN |
 |
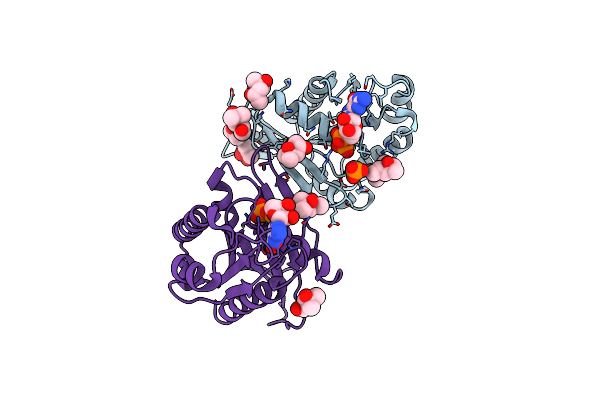
Organism: Myxococcus xanthus dk 1622
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-01-22 Classification: CELL CYCLE Ligands: MG, CTP, GOL |
 |
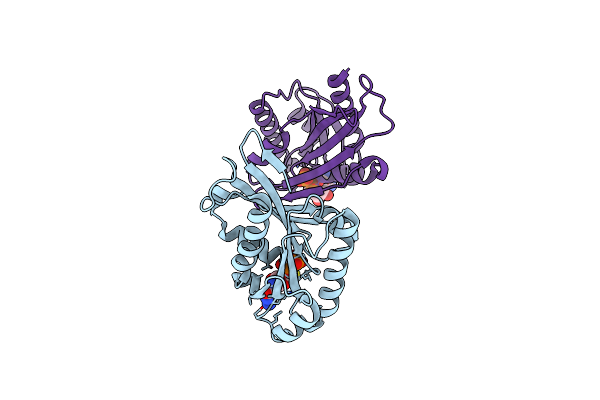
Myxococcus Xanthus Mgla Bound To Gdp And Pi With Mixed Inactive And Active Switch Region Conformations
Organism: Myxococcus xanthus dk 1622
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-12-04 Classification: CYTOSOLIC PROTEIN Ligands: GDP, PO4, MPD |
 |
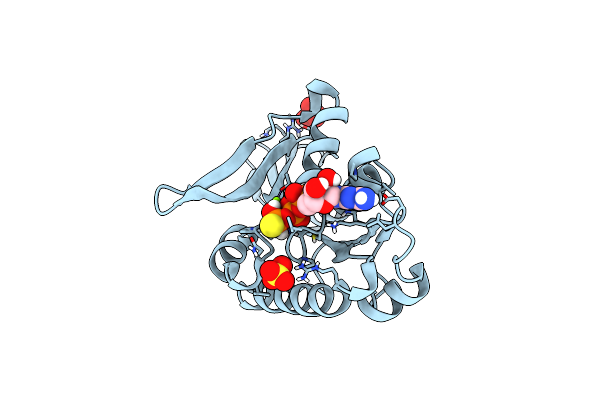
Organism: Myxococcus xanthus dk 1622
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2019-12-04 Classification: CYTOSOLIC PROTEIN Ligands: GDP |
 |
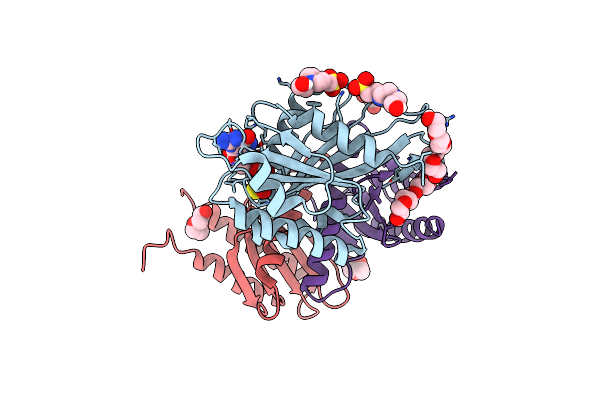
Organism: Myxococcus xanthus dk 1622
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2019-11-27 Classification: CYTOSOLIC PROTEIN Ligands: MG, GSP, SO4 |
 |
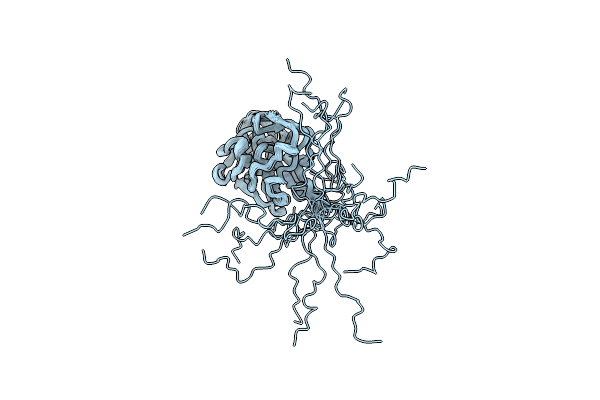
Organism: Myxococcus xanthus dk 1622, Myxococcus xanthus
Method: X-RAY DIFFRACTION Release Date: 2019-11-27 Classification: CYTOSOLIC PROTEIN Ligands: GSP, MG, EPE, 1PE, PEG |
 |
Organism: Myxococcus xanthus dk 1622
Method: SOLUTION NMR Release Date: 2019-06-19 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Aibr In Complex With Isovaleryl Coenzyme A And Operator Dna
Organism: Myxococcus xanthus dk 1622
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2016-12-28 Classification: TRANSCRIPTION Ligands: IVC |
 |
Crystal Structure Of Aibr In Complex With The Effector Molecule Isovaleryl Coenzyme A
Organism: Myxococcus xanthus dk 1622
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2016-12-21 Classification: TRANSCRIPTION Ligands: NI, CL, IVC |
 |
Organism: Myxococcus xanthus dk 1622
Method: ELECTRON MICROSCOPY Release Date: 2016-03-16 Classification: MOTOR PROTEIN |

