Search Count: 25
 |
Organism: Homo sapiens
Method: NEUTRON DIFFRACTION Resolution:2.30 Å Release Date: 2025-03-12 Classification: OXIDOREDUCTASE Ligands: MN, PEO |
 |
Organism: Homo sapiens
Method: NEUTRON DIFFRACTION Resolution:2.50 Å Release Date: 2025-03-12 Classification: OXIDOREDUCTASE Ligands: MN |
 |
Organism: Homo sapiens
Method: NEUTRON DIFFRACTION Resolution:2.28 Å Release Date: 2025-03-12 Classification: OXIDOREDUCTASE Ligands: MN |
 |
X-Ray Counterpart To The Neutron Structure Of Peroxide-Soaked Tyr34Phe Mnsod
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-03-12 Classification: OXIDOREDUCTASE Ligands: MN, PEO, PO4, K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-03-12 Classification: OXIDOREDUCTASE Ligands: MN, PO4, K |
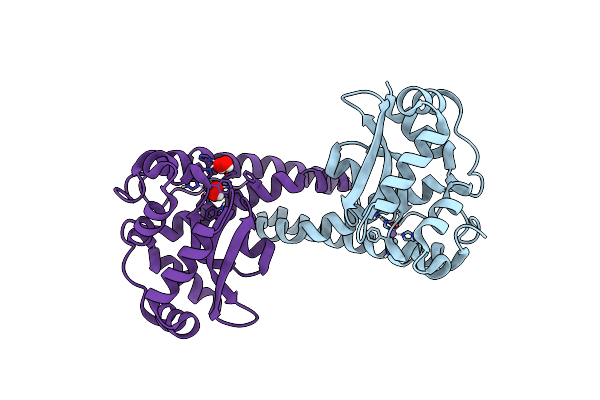 |
Organism: Homo sapiens
Method: NEUTRON DIFFRACTION Resolution:2.30 Å Release Date: 2024-07-31 Classification: OXIDOREDUCTASE Ligands: MN, PEO |
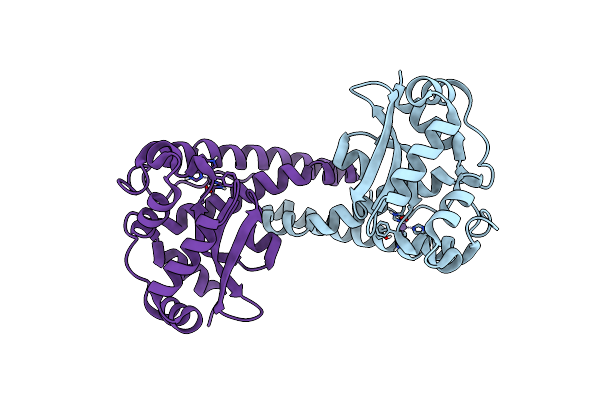 |
Organism: Homo sapiens
Method: NEUTRON DIFFRACTION Resolution:2.30 Å Release Date: 2024-07-31 Classification: OXIDOREDUCTASE Ligands: MN |
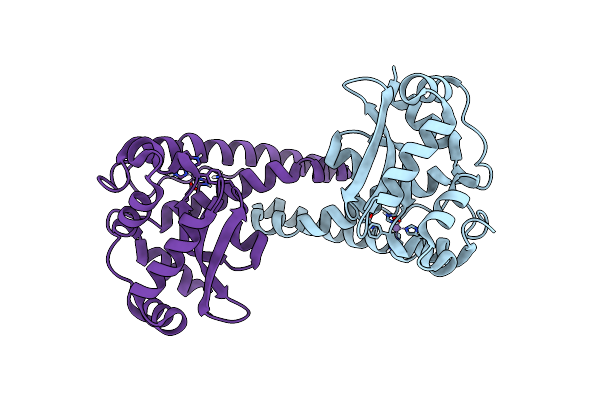 |
Organism: Homo sapiens
Method: NEUTRON DIFFRACTION Resolution:2.30 Å Release Date: 2024-07-31 Classification: OXIDOREDUCTASE Ligands: MN |
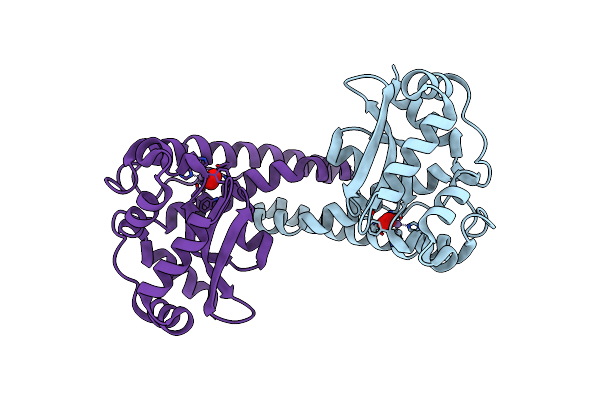 |
X-Ray Counterpart To The Neutron Structure Of Peroxide-Soaked Trp161Phe Mnsod
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-07-31 Classification: OXIDOREDUCTASE Ligands: MN, PEO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-07-31 Classification: OXIDOREDUCTASE Ligands: MN, PEO, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-07-31 Classification: OXIDOREDUCTASE Ligands: MN, PO4, K |
 |
X-Ray Structure Of The Fenna-Matthews-Olsen Antenna Complex From Prosthecochloris Aestuarii
Organism: Prosthecochloris aestuarii
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2019-03-13 Classification: PHOTOSYNTHESIS Ligands: BCL, SO4 |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-09-19 Classification: SUGAR BINDING PROTEIN Ligands: MG |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-09-19 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-09-19 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-09-19 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-09-19 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-12-27 Classification: OXIDOREDUCTASE Ligands: FOL, IPA, CA, CL |
 |
Neutron Crystallographic Structure Of Perdeuterated T4 Lysozyme Cysteine-Free Pseudo-Wild Type At Cryogenic Temperature
Organism: Enterobacteria phage t4
Method: NEUTRON DIFFRACTION Resolution:2.20 Å Release Date: 2017-07-26 Classification: HYDROLASE Ligands: CL, DOD |
 |
X-Ray Structure Of Perdeuterated T4 Lysozyme Cysteine-Free Pseudo-Wild Type At Cryogenic Temperature
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2017-07-26 Classification: HYDROLASE Ligands: PO4, NA, HED, CL, GOL |

