Search Count: 485
 |
Solution Structure Of An Uncharacterized Protein Containing A Duf1893 Domain From Borrelia Burgdorferi, A Pathogen Responsible For Lyme Disease. Seattle Structural Genomics Center For Infectious Disease Target Bobua.17726.A.A1.
Organism: Borreliella burgdorferi b31
Method: SOLUTION NMR Release Date: 2024-12-04 Classification: UNKNOWN FUNCTION |
 |
Solution Structure Of The Translation Initiation Factor If-1 From Neisseria Gonorrhoeae (Nccp11945). Seattle Structural Genomics Center For Infectious Disease Target Negoa.17902.A
Organism: Neisseria gonorrhoeae (strain atcc 700825 / fa 1090)
Method: SOLUTION NMR Release Date: 2024-10-16 Classification: RNA BINDING PROTEIN |
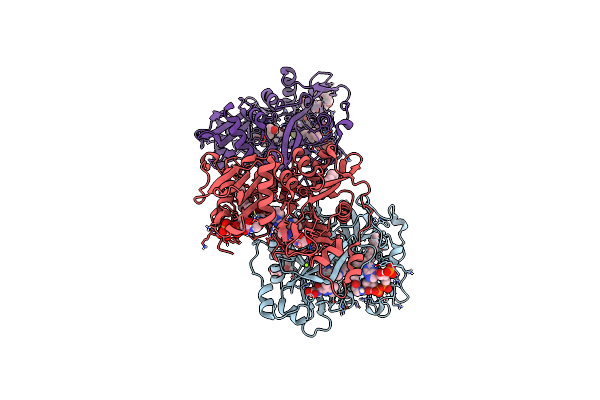 |
Crystal Structure Of Plasmodium Vivax Glycylpeptide N-Tetradecanoyltransferase (N-Myristoyltransferase, Nmt) Bound To Myristoyl-Coa And Inhibitor 9C
Organism: Plasmodium vivax sal-1
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-07-17 Classification: TRANSFERASE Ligands: MYA, A1AB7, CL, GOL, 1PE, MG, P33 |
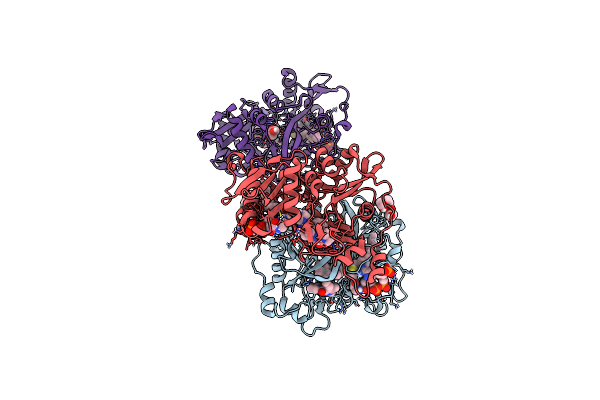 |
Crystal Structure Of Plasmodium Vivax Glycylpeptide N-Tetradecanoyltransferase (N-Myristoyltransferase, Nmt) Bound To Myristoyl-Coa And Inhibitor 10B
Organism: Plasmodium vivax sal-1
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2024-07-17 Classification: TRANSFERASE Ligands: MYA, A1AB8, CL, GOL, 1PE, P6G |
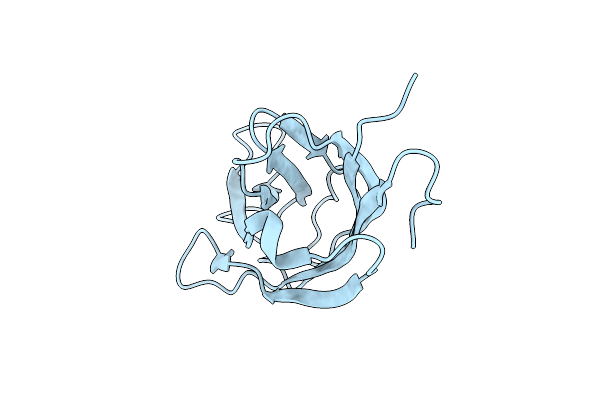 |
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2024-05-08 Classification: LIPID BINDING PROTEIN |
 |
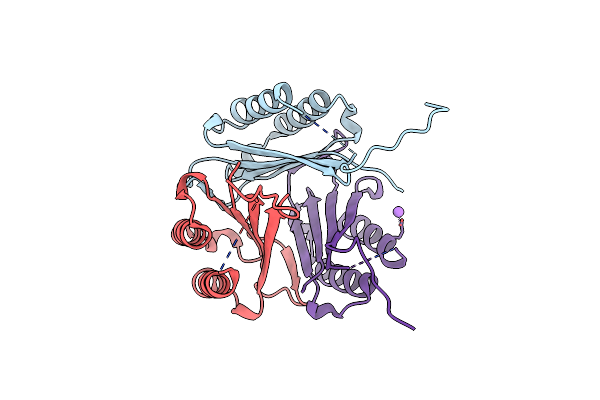
Glutamine Synthetase From Pseudomonas Aeruginosa, Filament Double-Unit In Compressed Conformation
Organism: Pseudomonas aeruginosa pao1
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: LIGASE |
 |
Crystal Structure Of Macrophage Migration Inhibitory Factor-1 (Mif1) From Onchocerca Volvulus
Organism: Onchocerca volvulus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-17 Classification: CYTOKINE |
 |
Crystal Structure Of Glutamyl-Trna Synthetase Glurs From Pseudomonas Aeruginosa (Zinc Bound)
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-12-27 Classification: LIGASE Ligands: FLC, SO4, ZN, 2PE, GOL |
 |
Crystal Structure Of Plasmodium Vivax Glycylpeptide N-Tetradecanoyltransferase (N-Myristoyltransferase, Nmt) Bound To Myristoyl-Coa And Inhibitor 12B
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-10-18 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, XOQ, GOL, SO4, CL, ACT, PEG, 1PE, PG4, PGE |
 |
Solution Structure For A Putative Type I Site-Specific Deoxyribonuclease From Neisseria Gonorrhoeae (Nccp11945). Seattle Structural Genomics Center For Infectious Disease Target Negoa.19201.A
Organism: Neisseria gonorrhoeae
Method: SOLUTION NMR Release Date: 2023-10-04 Classification: STRUCTURAL GENOMICS |
 |
Crystal Structure Of Glycine--Trna Ligase From Mycobacterium Tuberculosis (G5A Bound)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2023-09-13 Classification: TRANSFERASE Ligands: G5A, IMD, MES, EDO, MG, CL |
 |
Crystal Structure Of Glycine--Trna Ligase From Mycobacterium Tuberculosis (Amp-Mg Bound)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2023-06-28 Classification: LIGASE Ligands: MG, CA, EDO, GOL, IMD, AMP |
 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Inhibitor 1
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-05-31 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, XOF, PEG, CL, PG4, GOL |
 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-05-31 Classification: TRANSFERASE Ligands: MYA, EDO, PEG, GOL |
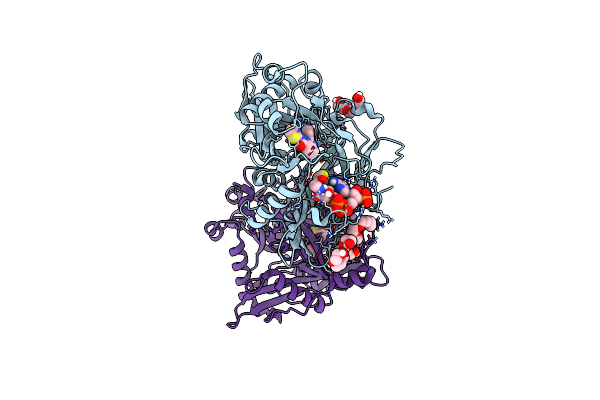 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Compound-2
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-05-31 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, XOL, PG4, 1PE, PGE, EDO, CL |
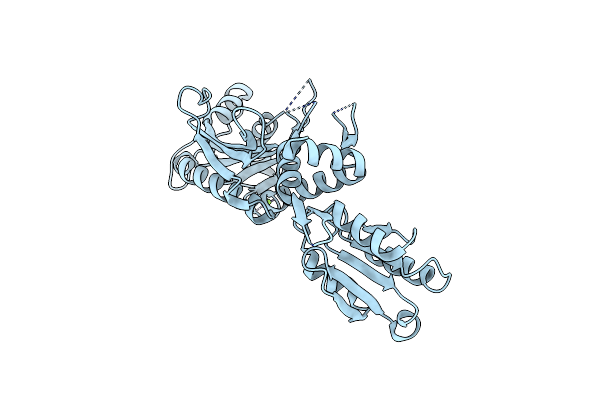 |
Crystal Structure Of Glycine Trna Ligase From Mycobacterium Thermoresistibile (Apo)
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: X-RAY DIFFRACTION Release Date: 2023-05-03 Classification: LIGASE Ligands: MG |
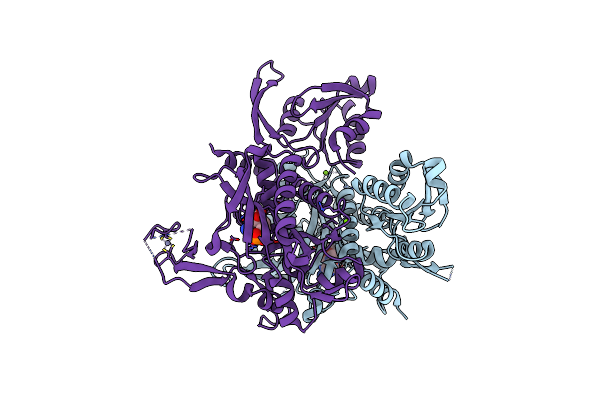 |
Crystal Structure Of Glycine Trna Ligase From Mycobacterium Thermoresistibile (Amp Bound)
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: X-RAY DIFFRACTION Release Date: 2023-05-03 Classification: LIGASE Ligands: AMP, MG, ZN |
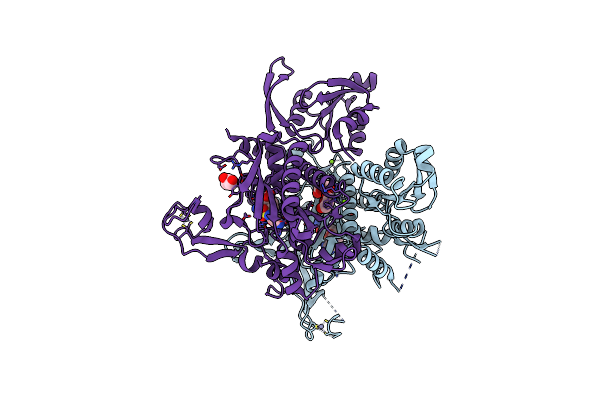 |
Crystal Structure Of Glycine Trna Ligase From Mycobacterium Thermoresistibile (Glycyl Adenylate Bound)
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: X-RAY DIFFRACTION Release Date: 2023-05-03 Classification: LIGASE Ligands: G5A, ZN, MG, GOL |
 |
Crystal Structure Of Dihydropteridine Reductase/Oxygen-Insensitive Nad(P)H Nitroreductase From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2022-07-27 Classification: OXIDOREDUCTASE Ligands: FMN, PO4, GOL |
 |
Solution Structure For Bartonella Henselae Bame, A Component Of The Beta-Barrel Assembly Machinery Complex. Seattle Structural Genomics Center For Infectious Disease Target Bahea.17605.A
Organism: Bartonella henselae str. houston-1
Method: SOLUTION NMR Release Date: 2022-03-02 Classification: PROTEIN BINDING |

