Search Count: 67
 |
D-2-Hydroxyacid Dehydrogenase (D2Hdh) From Haloferax Mediterranei In Complex With Potassium, 2-Ketohexanoic Acid, Nadp+ And Chloride
Organism: Haloferax mediterranei
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: NAP, 7N5, K, MG, CL |
 |
D-2-Hydroxyacid Dehydrogenase (D2-Hdh) From Haloferax Mediterranei Apo-Enzyme (2.25 A Resolution)
Organism: Haloferax mediterranei
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: MG, NA |
 |
D-2-Hydroxyacid Dehydrogenase (D2Hdh) From Haloferax Mediterranei In Complex With 2-Ketohexanoic Acid, Nad+ And Chloride (1.16 A Resolution)
Organism: Haloferax mediterranei
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: NAD, 7N5, ACT, MG, NA, CL |
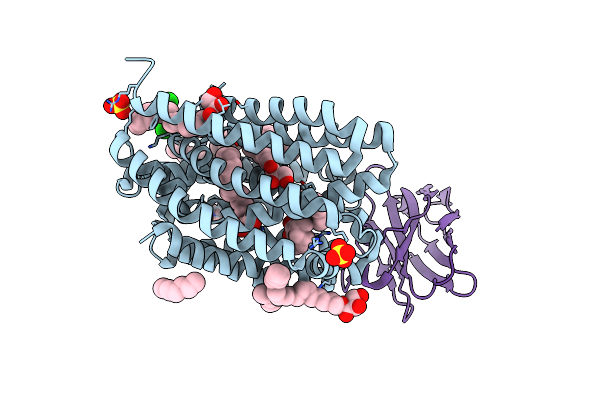 |
Lipid Iii Flippase Wzxe With Nb10 Nanobody In Outward-Facing Conformation At 2.7552 A
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: MPG, OCT, SO4, ZN, CL |
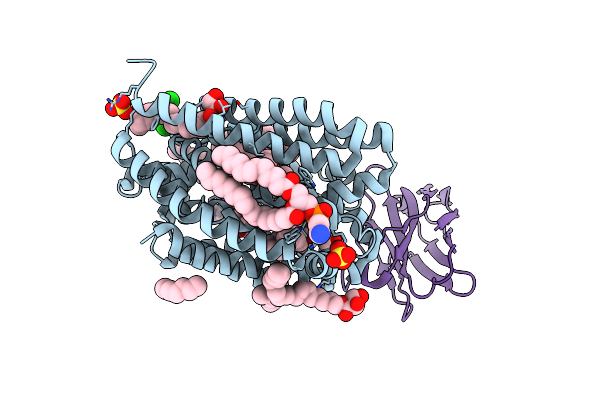 |
Lipid Iii Flippase Wzxe With Nb10 Nanobody In Outward-Facing Conformation At 0.9688 A
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: MPG, LOP, OCT, CL, SO4, ZN |
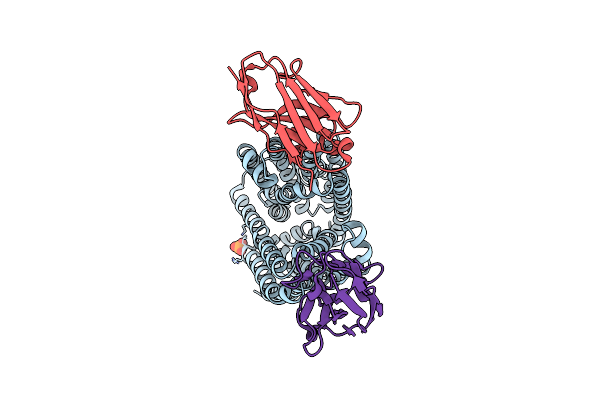 |
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Outward-Facing Conformation - Crystal 1
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: PO4 |
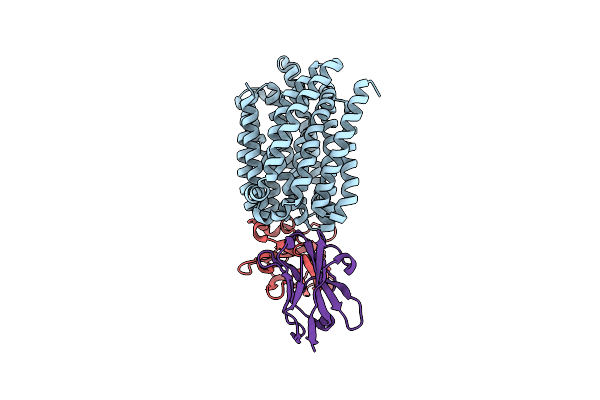 |
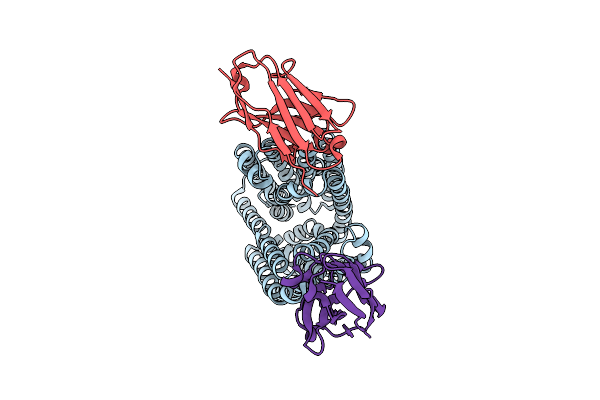
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Inward-Facing Conformation - Crystal 1
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
 |
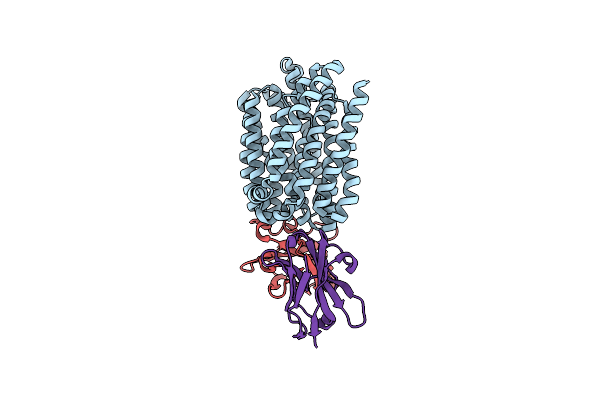
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Outward-Facing Conformation - Crystal 2
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
 |
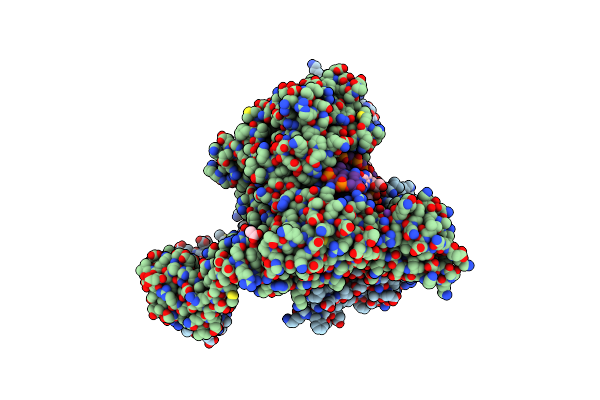
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Inward-Facing Conformation - Crystal 2
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
 |
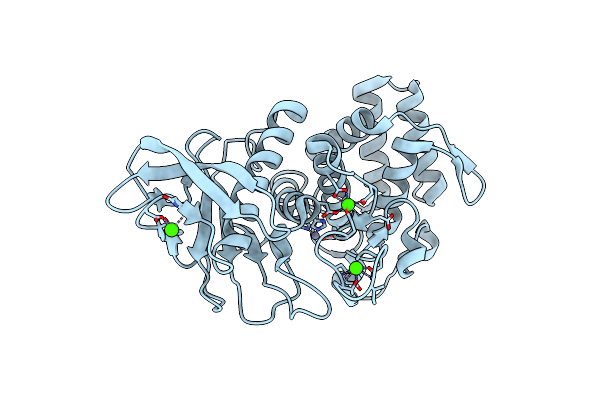
Experimental Localization Of Metal-Binding Sites Reveals The Role Of Metal Ions In The Delafloxacin-Stabilized Streptococcus Pneumoniae Topoisomerase Iv Dna Cleavage Complex
Organism: Streptococcus pneumoniae, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2024-10-16 Classification: DNA BINDING PROTEIN Ligands: MPD, MG, K, CL, ACT, TE9 |
 |
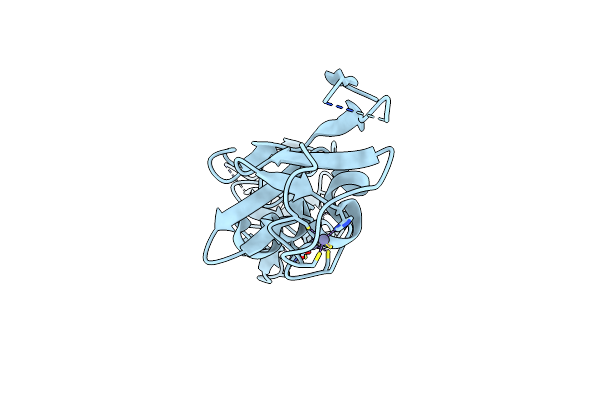
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-10-02 Classification: HYDROLASE Ligands: CA, ZN |
 |
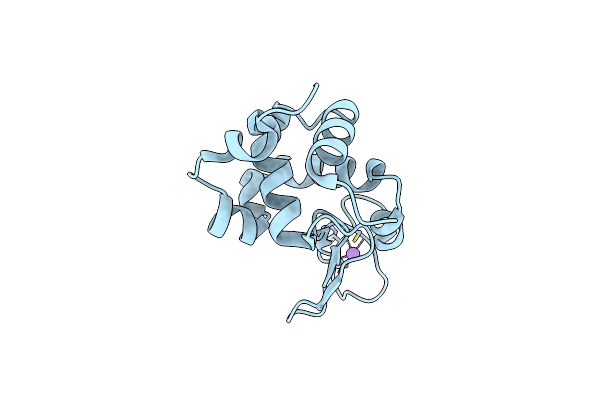
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-10-02 Classification: HYDROLASE Ligands: ZN |
 |
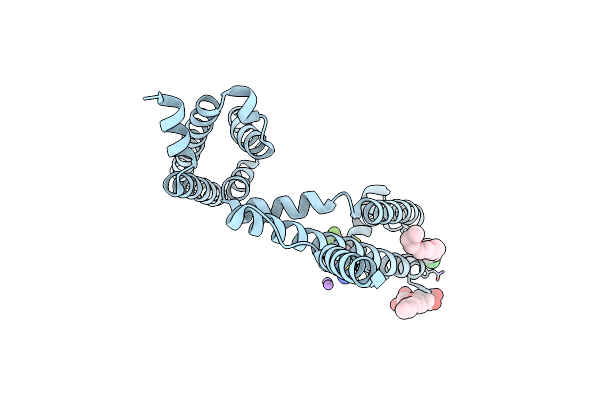
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-10-02 Classification: HYDROLASE |
 |
Organism: Magnetococcus marinus mc-1
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-09-18 Classification: MEMBRANE PROTEIN |
 |
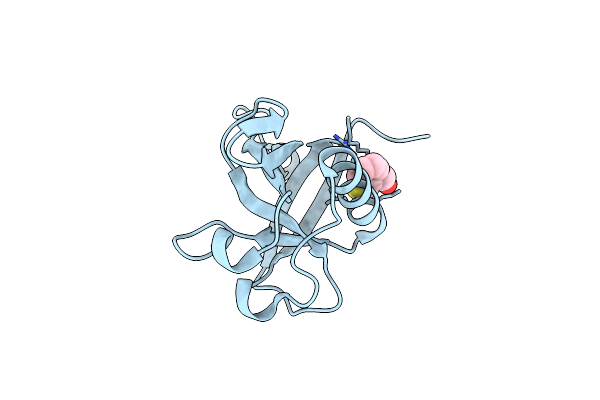
Crystal Structure Of N-Terminal Sars-Cov-2 Nsp1 In Complex With Fragment Hit 1E7 Refined Against The Anomalous Diffraction Data
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2024-06-19 Classification: PROTEIN BINDING Ligands: A1H0G |
 |
Crystal Structure Of N-Terminal Sars-Cov-2 Nsp1 In Complex With Fragment Hit 7G3 Refined Against The Anomalous Diffraction Data
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2024-06-19 Classification: PROTEIN BINDING Ligands: A1H0L |
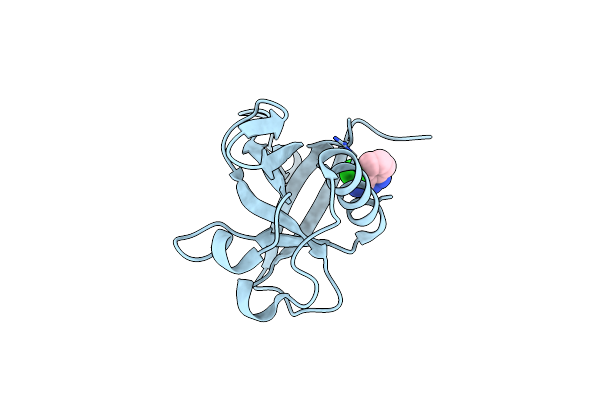 |
Crystal Structure Of N-Terminal Sars-Cov-2 Nsp1 In Complex With Fragment Hit 9D4 Refined Against The Anomalous Diffraction Data
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2024-06-19 Classification: PROTEIN BINDING Ligands: EQT |
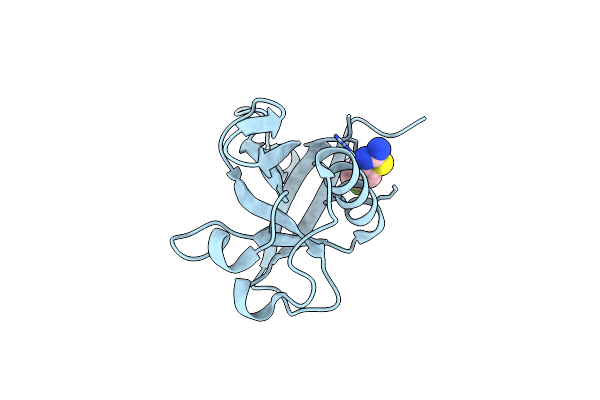 |
Crystal Structure Of N-Terminal Sars-Cov-2 Nsp1 In Complex With Fragment Hit 11A7 Refined Against The Anomalous Diffraction Data
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2024-06-19 Classification: PROTEIN BINDING Ligands: FBB |
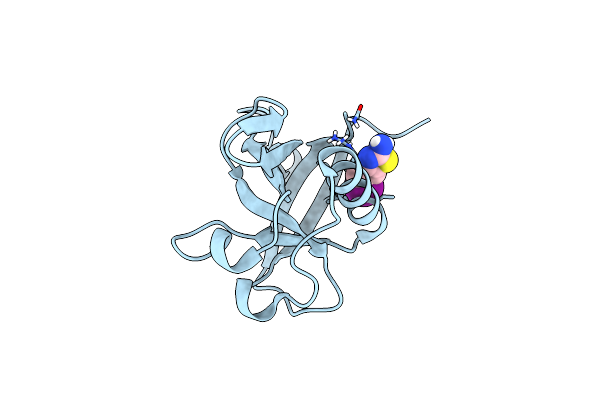 |
Crystal Structure Of N-Terminal Sars-Cov-2 Nsp1 In Complex With Fragment Hit 11A7_Al5 Refined Against The Anomalous Diffraction Data
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2024-06-19 Classification: PROTEIN BINDING Ligands: A1H0K |
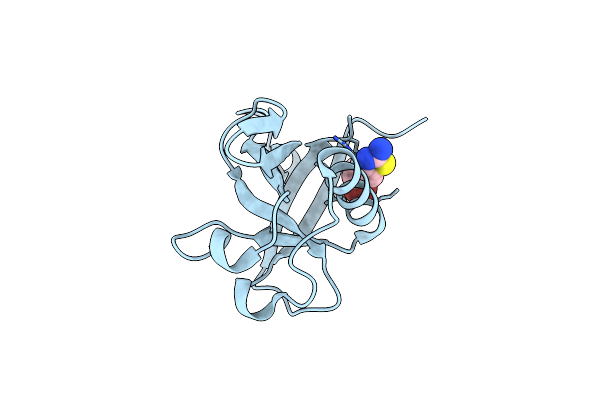 |
Crystal Structure Of N-Terminal Sars-Cov-2 Nsp1 In Complex With Fragment Hit 11A7_Al6 Refined Against The Anomalous Diffraction Data
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2024-06-19 Classification: PROTEIN BINDING Ligands: A1H0M |

