Search Count: 21
 |
Crystal Structure Of The N-Terminal Domain Of Fatty Acid Kinase A (Faka) From Staphylococcus Aureus (Mn And Amp-Pnp)
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-11-06 Classification: LIGASE Ligands: ANP, MN |
 |
Crystal Structure Of The N-Terminal Domain Of Fatty Acid Kinase A (Faka) From Staphylococcus Aureus (Mg And Amp-Pnp)
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2024-11-06 Classification: LIGASE Ligands: ANP, MG |
 |
Crystal Structure Of The N-Terminal Domain Of Fatty Acid Kinase A (Faka) From Staphylococcus Aureus (Apo)
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-11-06 Classification: LIGASE |
 |
Crystal Structure Of The N-Terminal Domain Of Fatty Acid Kinase A (Faka) From Staphylococcus Aureus (Mg And Adp)
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-11-06 Classification: LIGASE Ligands: ADP, MG, PG4 |
 |
Exploiting Activation And Inactivation Mechanisms In Type I-C Crispr-Cas3 For Genome Editing Applications
Organism: Neisseria lactamica, Rhodobacter phage rcnl1
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: HYDROLASE/RNA |
 |
Exploiting Activation And Inactivation Mechanisms In Type I-C Crispr-Cas3 For Genome Editing Applications
Organism: Rhodobacter phage rcnl1, Neisseria lactamica
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: HYDROLASE/RNA |
 |
Exploiting Activation And Inactivation Mechanisms In Type I-C Crispr-Cas3 For Genome Editing Applications
Organism: Neisseria lactamica
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: DNA BINDING PROTEIN Ligands: MG, PO4 |
 |
Exploiting Activation And Inactivation Mechanisms In Type I-C Crispr-Cas3 For Genome Editing Applications
Organism: Neisseria lactamica
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: HYDROLASE/RNA |
 |
Exploiting Activation And Inactivation Mechanisms In Type I-C Crispr-Cas3 For Genome Editing Applications
Organism: Neisseria lactamica
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: HYDROLASE/RNA/DNA |
 |
Exploiting Activation And Inactivation Mechanisms In Type I-C Crispr-Cas3 For Genome Editing Applications
Organism: Neisseria lactamica
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: HYDROLASE/RNA/DNA |
 |
Fabp5 In Complex With 6-Chloro-4-Phenyl-2-Piperidin-1-Yl-3-(1H-Tetrazol-5-Yl)-Quinoline
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-01-25 Classification: LIPID BINDING PROTEIN Ligands: DMS, SO4, 65X |
 |
Fabp4 In Complex With 6-Chloro-2-Isopropyl-4-(3-Isopropyl-Phenyl)-Quinoline-3-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2016-12-14 Classification: TRANSPORT PROTEIN Ligands: 65Y |
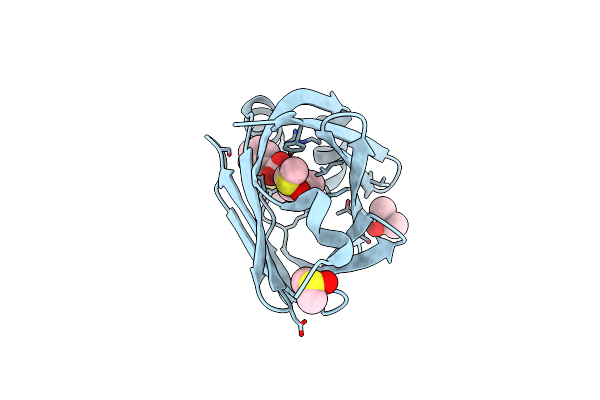 |
Fabp4_3 In Complex With 6,8-Dichloro-4-Phenyl-2-Piperidin-1-Yl-Quinoline-3-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2016-12-14 Classification: TRANSPORT PROTEIN Ligands: 65Z, DMS |
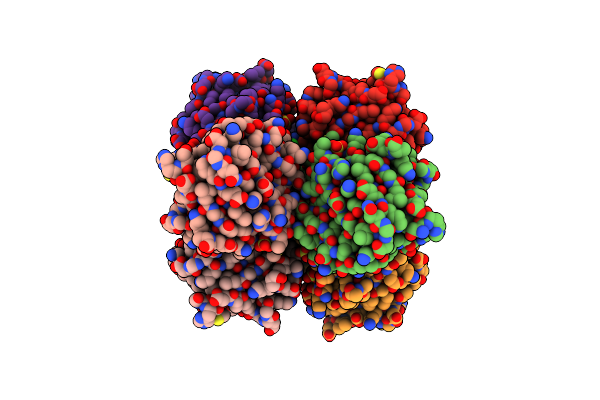 |
Human Fabp3 In Complex With 6-Chloro-2-Methyl-4-Phenyl-Quinoline-3-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-12-14 Classification: LIPID BINDING PROTEIN Ligands: SO4, 5M8, CL |
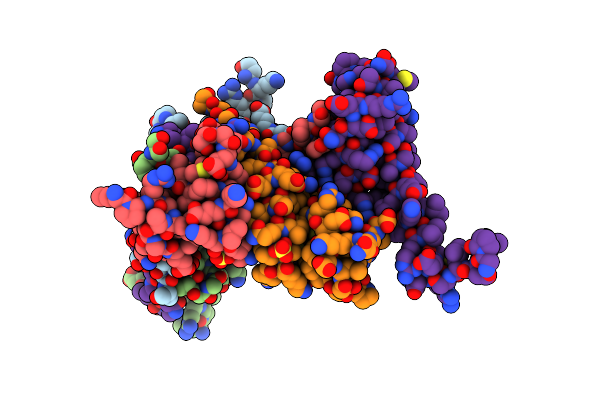 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-05-23 Classification: TRANSCRIPTION Ligands: SO4 |
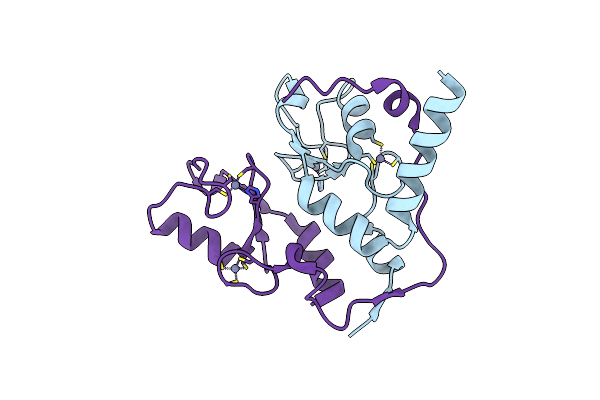 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-05-23 Classification: METAL BINDING PROTEIN/LIGASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-06-12 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2003-06-12 Classification: HYDROLASE |
 |
Crystal Structure Of Protein Tyrosine Phosphatase 1B Complexed With A Tri-Phosphorylated Peptide (Rdi(Ptr)Etd(Ptr)(Ptr)Rk) From The Insulin Receptor Kinase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-01-17 Classification: HYDROLASE, SIGNALING PROTEIN |
 |
Crystal Structure Of Protein Tyrosine Phosphatase 1B Complexed With A Mono-Phosphorylated Peptide (Etdy(Ptr)Rkggkgll) From The Insulin Receptor Kinase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2001-01-17 Classification: HYDROLASE, SIGNALING PROTEIN |

