Search Count: 16
 |
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2020-09-09 Classification: MEMBRANE PROTEIN Ligands: MC3 |
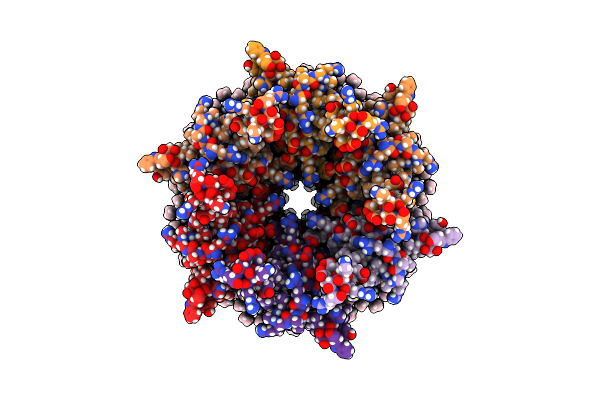 |
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2020-09-09 Classification: MEMBRANE PROTEIN Ligands: MC3 |
 |
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2020-09-09 Classification: MEMBRANE PROTEIN Ligands: MC3 |
 |
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2020-09-09 Classification: MEMBRANE PROTEIN Ligands: MC3 |
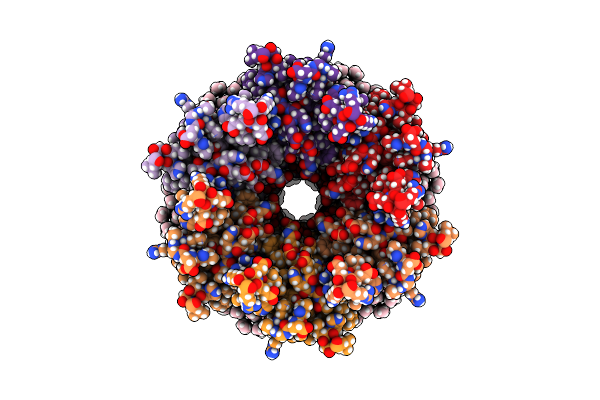 |
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2020-09-09 Classification: MEMBRANE PROTEIN Ligands: MC3 |
 |
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2020-09-09 Classification: MEMBRANE PROTEIN Ligands: MC3 |
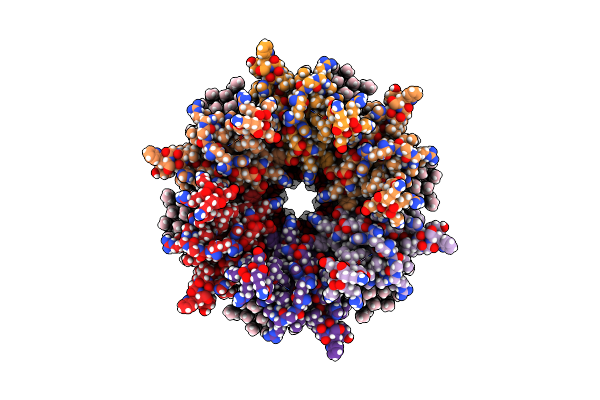 |
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2020-09-09 Classification: MEMBRANE PROTEIN Ligands: MC3 |
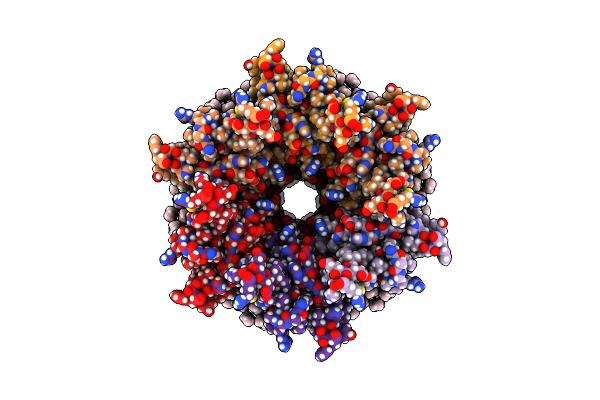 |
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2020-09-09 Classification: MEMBRANE PROTEIN Ligands: MC3 |
 |
Structure Of Connexin-46 Intercellular Gap Junction Channel At 3.4 Angstrom Resolution By Cryoem
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2018-12-12 Classification: MEMBRANE PROTEIN |
 |
Structure Of Connexin-50 Intercellular Gap Junction Channel At 3.4 Angstrom Resolution By Cryoem
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2018-12-12 Classification: MEMBRANE PROTEIN |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2017-06-21 Classification: TRANSFERASE |
 |
Crystal Structure Of Methionyl-Trna Synthetase Metrs From Brucella Melitensis In Complex With Inhibitor Chem 1312
Organism: Brucella suis biovar 1 (strain 1330)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2016-05-25 Classification: ligase/ligase inhibitor Ligands: 0OU |
 |
Crystal Structure Of Methionyl-Trna Synthetase Metrs From Brucella Melitensis In Complex With Inhibitor Chem 1415
Organism: Brucella suis biovar 1 (strain 1330)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-05-25 Classification: LIGASE Ligands: 415, EDO |
 |
Crystal Structure Of 2-C-Methyl-D-Erythritol 4-Phosphate Cytidylyltransferase (Ispd) From Burkholderia Thailandensis Complexed With Ctp
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-04-29 Classification: TRANSFERASE Ligands: CTP, MG, ACT, CAD, GOL |
 |
Crystal Structure Of Methionyl-Trna Synthetase Metrs From Brucella Melitensis In Complex With Inhibitor 1-{3-[(3,5-Dichlorobenzyl)Amino]Propyl}-3-Thiophen-3-Ylurea
Organism: Brucella melitensis
Method: X-RAY DIFFRACTION Release Date: 2015-04-15 Classification: ligase/ligase inhibitor Ligands: 43E, EDO |
 |
Crystal Structure Of Methionyl-Trna Synthetase Metrs From Brucella Melitensis Bound To Selenomethionine
Organism: Brucella melitensis biovar abortus 2308
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2012-02-15 Classification: LIGASE Ligands: MSE |

