Search Count: 18
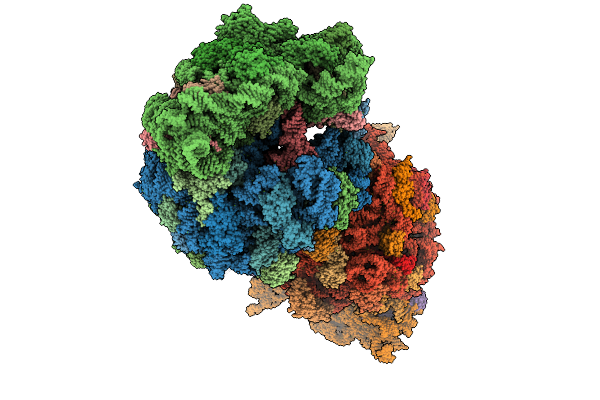 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With O-Cresomycin, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.50A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: RIBOSOME Ligands: MG, K, A1A1F, ZN, SF4 |
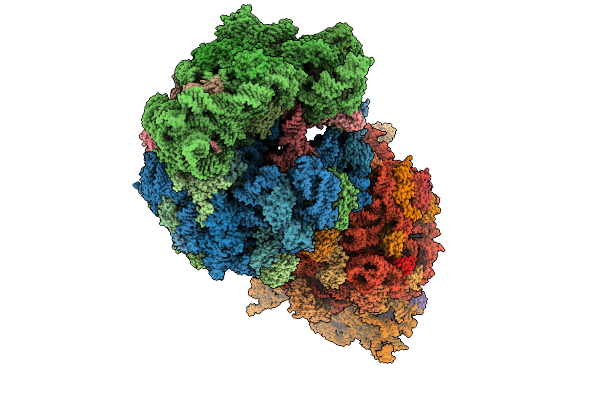 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With C-Cresomycin, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.50A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: RIBOSOME Ligands: MG, K, A1A1J, ZN, SF4 |
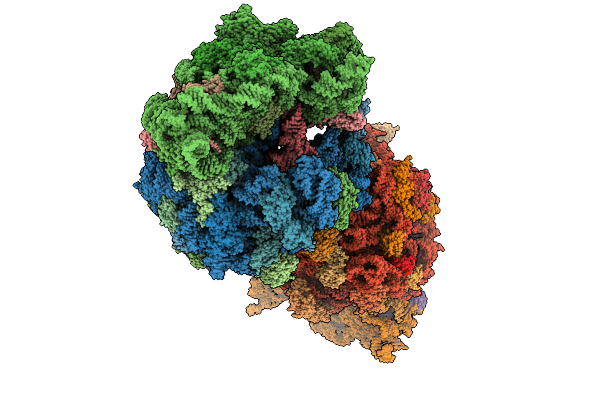 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Se-Cresomycin, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.45A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: RIBOSOME Ligands: MG, K, A1A1K, ZN, SF4 |
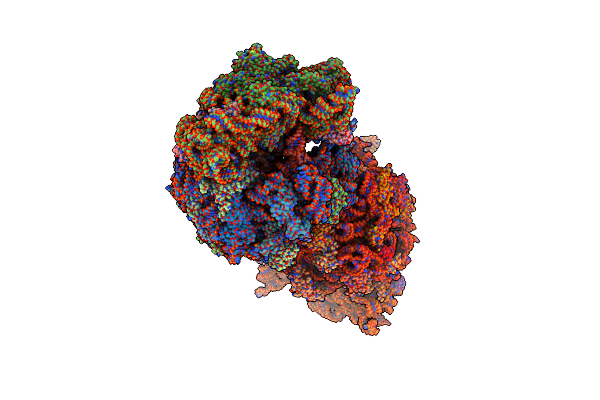 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Bt-33, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.50A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, A1A1L, ZN, SF4, K |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Cresomycin, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.70A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-02-21 Classification: Ribosome/RNA Ligands: MG, K, WC9, ZN, SF4 |
 |
Crystal Structure Of The A2058-N6-Dimethylated Thermus Thermophilus 70S Ribosome In Complex With Cresomycin, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.70A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2024-02-21 Classification: Ribosome/RNA Ligands: MG, WC9, ZN, SF4, K |
 |
Crystal Structure Of The A2503-C2,C8-Dimethylated Thermus Thermophilus 70S Ribosome In Complex With Cresomycin, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.70A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-02-21 Classification: Ribosome/RNA Ligands: MG, K, WC9, ZN, SF4 |
 |
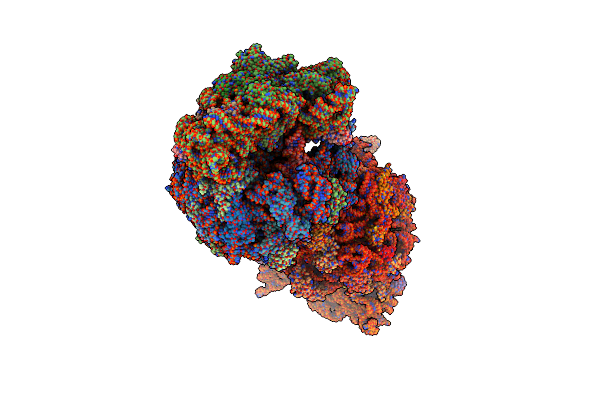
Crystal Structure Of The A2503-C2,C8-Dimethylated Thermus Thermophilus 70S Ribosome In Complex With Mrna, Aminoacylated A-Site Phe-Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.55A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2023-12-27 Classification: RIBOSOME Ligands: MG, K, ZN, SF4 |
 |
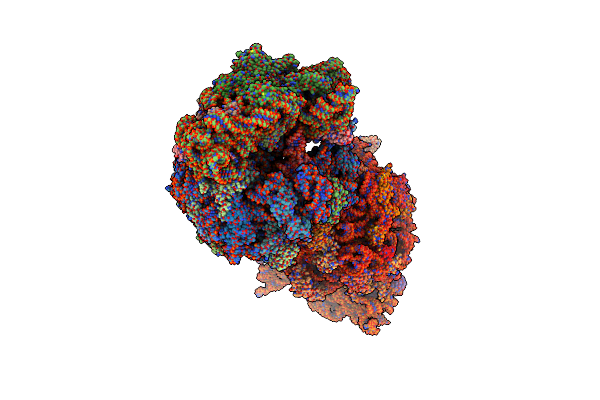
Crystal Structure Of The A2503-C2,C8-Dimethylated Thermus Thermophilus 70S Ribosome In Complex With Mrna, Aminoacylated A-Site Phe-Trnaphe, Peptidyl P-Site Fmthsmrc-Trnamet, And Deacylated E-Site Trnaphe At 2.45A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2023-12-27 Classification: RIBOSOME Ligands: MG, K, ZN, SF4 |
 |
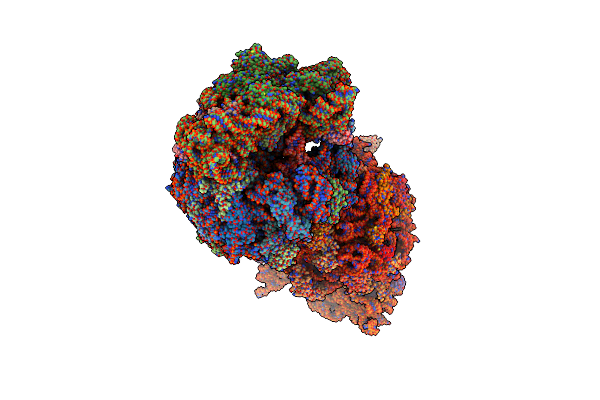
Crystal Structure Of The A2503-C2,C8-Dimethylated Thermus Thermophilus 70S Ribosome In Complex With Iboxamycin, Mrna, Deacylated A- And E-Site Trnaphe, And Aminoacylated P-Site Fmet-Trnamet At 2.55A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2023-12-27 Classification: RIBOSOME Ligands: MG, 6IF, ZN, SF4, K |
 |
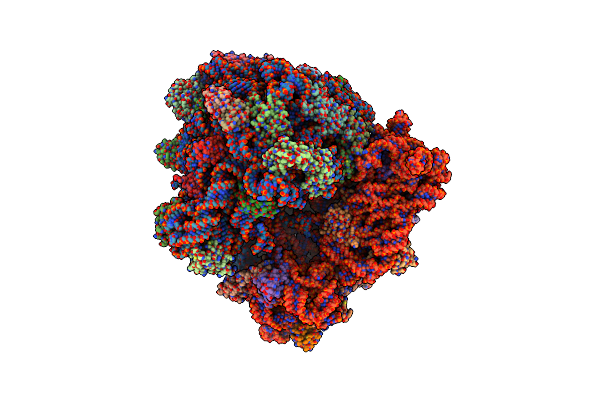
Crystal Structure Of The A2503-C2,C8-Dimethylated Thermus Thermophilus 70S Ribosome In Complex With Tylosin, Mrna, Aminoacylated A-Site Phe-Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.65A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-12-27 Classification: RIBOSOME Ligands: MG, K, TYK, ZN, SF4 |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Tylosin, Mrna, Deacylated A- And E-Site Trnaphe, And Deacylated P-Site Trnamet At 2.70A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-12-27 Classification: RIBOSOME Ligands: MG, TYK, ZN, SF4, K |
 |
Organism: Neobacillus vireti, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: RIBOSOME Ligands: K, MG, ZN, ATP |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Iboxamycin, Mrna, Deacylated A- And E-Site Trnas, And Aminoacylated P-Site Trna At 2.50A Resolution
Organism: Escherichia coli, Escherichia virus t4, Thermus thermophilus hb8, Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-10-13 Classification: RIBOSOME/RNA Ligands: MG, 6IF, ZN, SF4, K |
 |
Crystal Structure Of The A2058-Dimethylated Thermus Thermophilus 70S Ribosome In Complex With Iboxamycin, Mrna, Deacylated A- And E-Site Trnas, And Aminoacylated P-Site Trna At 2.60A Resolution
Organism: Escherichia coli, Escherichia virus t4, Thermus thermophilus hb8, Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-10-13 Classification: Ribosome/RNA Ligands: MG, K, 6IF, ZN, SF4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-09-23 Classification: TRANSFERASE Ligands: C1I, FMT |
 |
Method: X-RAY DIFFRACTION
Resolution:1.09 Å Release Date: 2013-10-23 Classification: ANTIBIOTIC Ligands: 18Y, GUN, DMS |
 |
Organism: Streptomyces carzinostaticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1993-10-31 Classification: ANTIBACTERIAL AND ANTITUMOR PROTEIN Ligands: MRD, CHR |

