Search Count: 25
 |
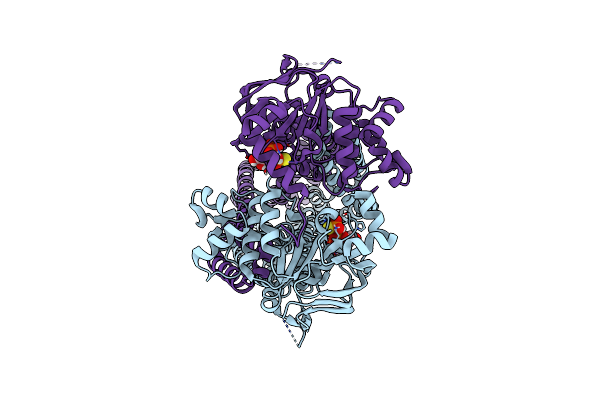
Cryo-Em Structure Of Irtab 2Xeq Mutant In Outward-Occluded State In Nanodisc In Complex With Mycobactin
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, ZN, A1IRY |
 |
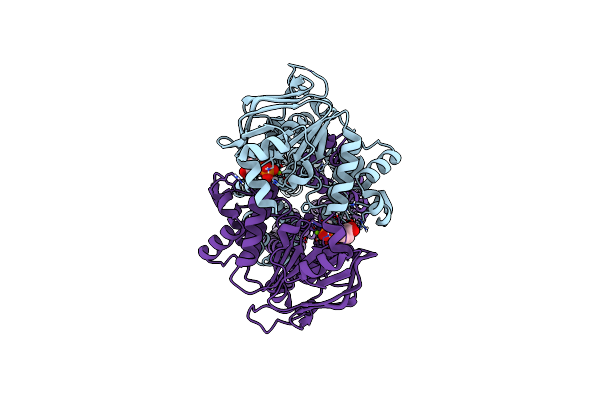
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: AGS |
 |
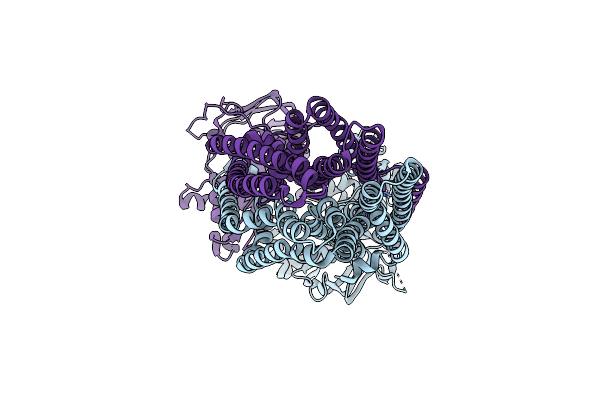
Cryo-Em Structure Of Irtab In Outward-Occluded State In Nanodisc In Complex With Adp-Vanadate
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: AOV, MG, ZN |
 |
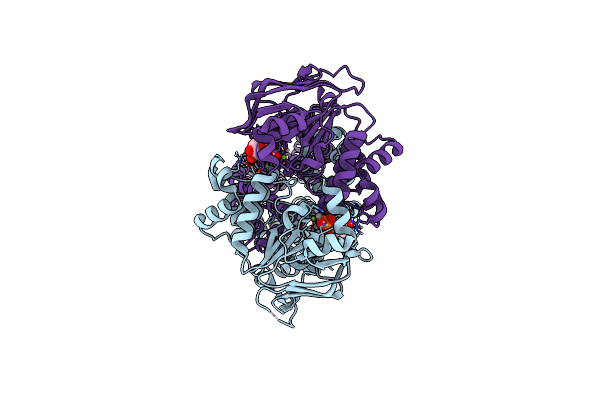
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: ZN |
 |
Cryo-Em Structure Of Irtab In Outward-Occluded State In Lmng In Complex With Adp-Vanadate
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: AOV, MG, ZN |
 |
Cryo-Em Structure Of Irtab 2Xeq Mutant In Outward-Occluded State In Nanodisc
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, ZN |
 |
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Cryo-Em Structure Of Irtab In Inward-Facing State In Presence Of Mycobactin Under Turnover Conditions In Lmng
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: ADP, ZN |
 |
Cryo-Em Structure Of Irtab In Outward-Occluded State In Presence Of Mycobactin Under Turnover Conditions In Lmng
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, ZN |
 |
Cryo-Em Structure Of Irtab In Inward-Facing State Under Turnover Conditions In Lmng
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: ADP |
 |
Cryo-Em Structure Of Irtab In Outward-Occluded State Under Turnover Conditions In Lmng
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, ZN |
 |
Cryo-Em Structure Of Irtab 3Xhtoa Mutant In Inward-Facing State In Presence Of Mycobactin Under Turnover Conditions In Lmng
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: ADP |
 |
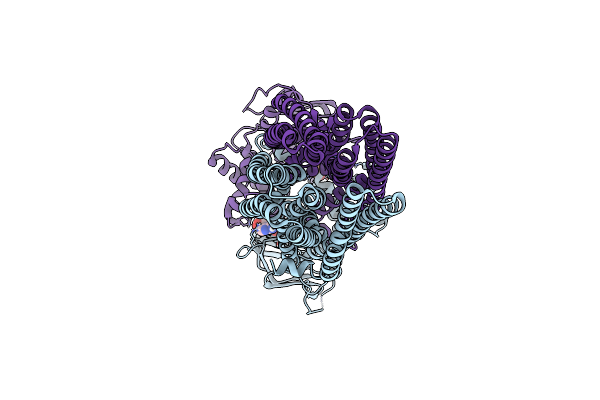
Cryo-Em Structure Of Irtab 3Xhtoa Mutant In Outward-Occluded State In Presence Of Mycobactin Under Turnover Conditions In Lmng
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: ATP |
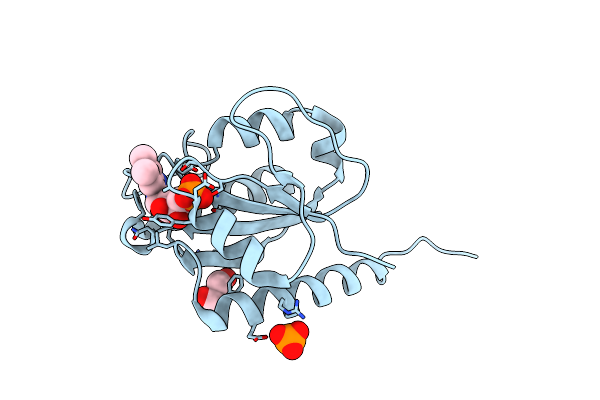 |
Structure Of Mycobacterium Thermoresistibile Nrdi(Oxidised) Determined At 1.1 Angstrom Resolution
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2024-10-09 Classification: FLAVOPROTEIN Ligands: FMN, PO4, GOL |
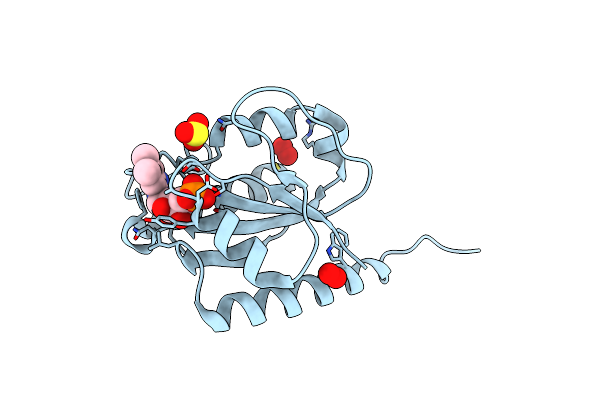 |
Structure Of Mycobacterium Thermoresistibile Nrdi(Reduced) Determined At 1.1 Angstrom Resolution
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2024-10-09 Classification: FLAVOPROTEIN Ligands: FMN, SO3, PEO, PER, CL |
 |
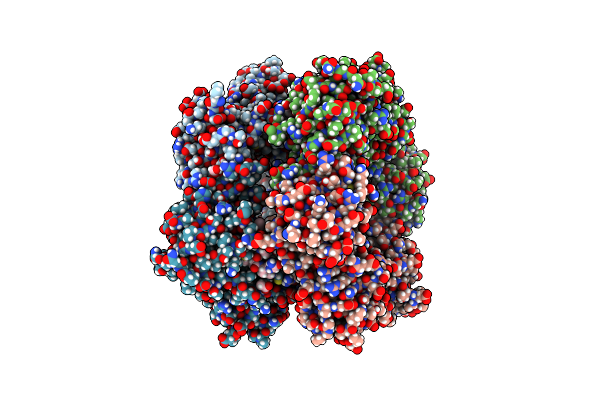
Crystal Structure Of Glycine--Trna Ligase Active Site Chimera From Mycobacterium Thermoresistibile/Tuberculosis (G5A Bound)
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2023-09-13 Classification: TRANSFERASE |
 |
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: FMT, ACT |
 |
Crystal Structure Of Glycine Trna Ligase From Mycobacterium Thermoresistibile (Apo)
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2023-05-03 Classification: LIGASE Ligands: MG |

