Search Count: 83
 |
Organism: Mycobacterium smegmatis str. mc2 155, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-02-19 Classification: REPLICATION/DNA Ligands: 0KX, MG |
 |
Organism: Mycobacterium smegmatis str. mc2 155, Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2020-02-19 Classification: REPLICATION/DNA Ligands: 0KX, MG |
 |
Crystal Structure At 1.3 Angstroms Resolution Of A Novel Udg, Udgx, From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4 |
 |
Organism: Mycolicibacterium smegmatis mc2 155, Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2019-01-23 Classification: HYDROLASE Ligands: ADP, MG, PO4 |
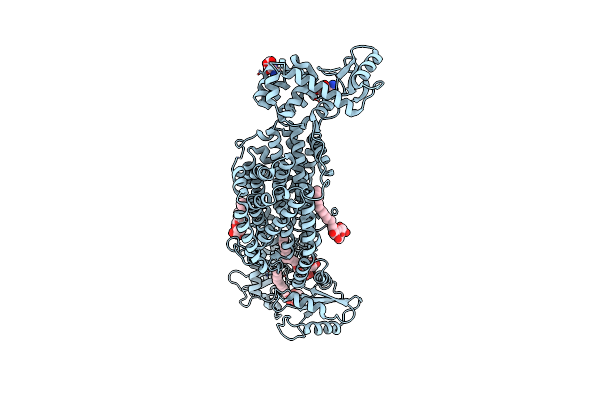 |
Crystal Structure Of Mycolic Acid Transporter Mmpl3 From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis str. mc2 155, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-12-26 Classification: MEMBRANE PROTEIN, HYDROLASE Ligands: MHA, L6T |
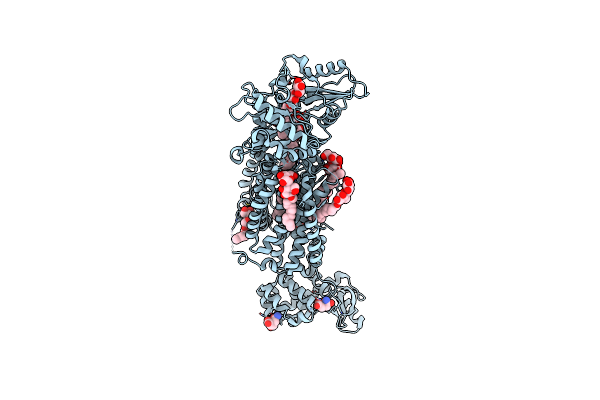 |
Crystal Structure Of Mycolic Acid Transporter Mmpl3 From Mycobacterium Smegmatis Complexed With Sq109
Organism: Mycobacterium smegmatis str. mc2 155, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-12-26 Classification: MEMBRANE PROTEIN, HYDROLASE Ligands: 3RX, LMT, MHA |
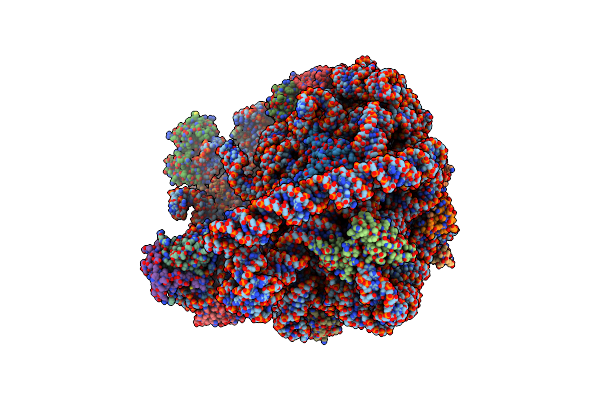 |
Cryo-Em Structure Of Mycobacterium Smegmatis C(Minus) 50S Ribosomal Subunit
Organism: Mycobacterium smegmatis str. mc2 155, Mycobacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: ELECTRON MICROSCOPY Release Date: 2018-10-03 Classification: RIBOSOME |
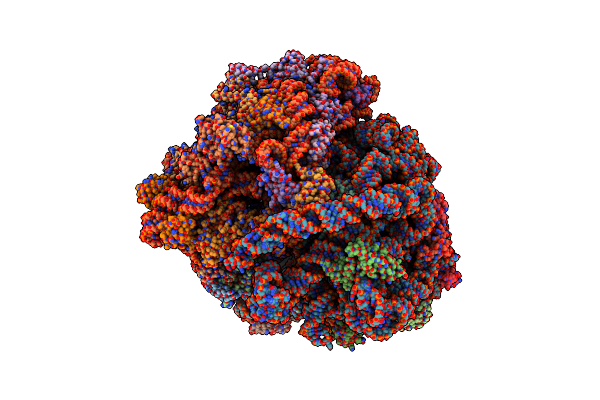 |
Organism: Mycobacterium smegmatis str. mc2 155, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2018-09-26 Classification: RIBOSOME |
 |
Organism: Mycobacterium smegmatis str. mc2 155, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2018-09-26 Classification: RIBOSOME |
 |
Organism: Mycobacterium smegmatis str. mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2018-09-26 Classification: RIBOSOME |
 |
Cryo-Em Structure Of Mycobacterium Smegmatis 70S C(Minus) Ribosome 70S-Mpy Complex
Organism: Mycobacterium smegmatis str. mc2 155, Mycobacterium smegmatis (strain atcc 700084 / mc(2)155), Mycobacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2018-09-26 Classification: RIBOSOME |
 |
Cryo-Em Structure Of Mycobacterium Smegmatis C(Minus) 30S Ribosomal Subunit With Mpy
Organism: Mycobacterium smegmatis str. mc2 155, Mycobacterium smegmatis (strain atcc 700084 / mc(2)155), Mycobacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2018-09-19 Classification: RIBOSOME |
 |
Crystal Structure Of A Mycobacterium Smegmatis Rna Polymerase Transcription Initiation Complex With Inhibitor Kanglemycin A
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155), Mycobacterium smegmatis str. mc2 155, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2018-08-15 Classification: transcription/dna/antibiotic Ligands: KNG, SO4, ZN, MG, GLU, EDO |
 |
Crystal Structure Of A Mycobacterium Smegmatis Rna Polymerase Transcription Initiation Complex With Inhibitor Rifampicin
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155), Mycobacterium smegmatis str. mc2 155, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2018-08-15 Classification: TRANSCRIPTION Ligands: SO4, EDO, RFP, ZN, GLU |
 |
Crystal Structure Of N-Acetyl-D-Glucosamine-6-Phosphate Deacetylase From Mycobacterium Smegmatis.
Organism: mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2018-05-16 Classification: HYDROLASE Ligands: ZN |
 |
Structural Of Start Superfamily Protein Msmeg_0129 From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-03-21 Classification: UNKNOWN FUNCTION |
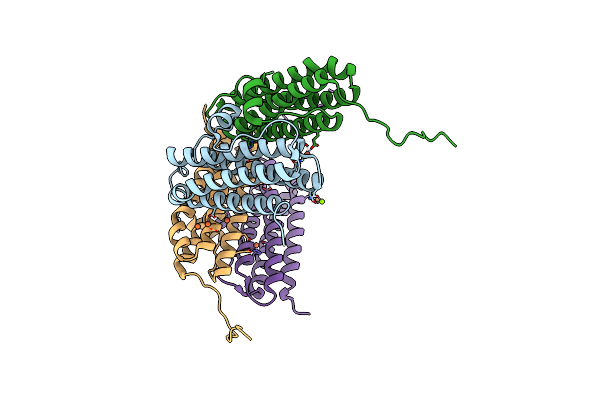 |
Crystal Structure Of The Second Dna-Binding Protein Under Starvation From Mycobacterium Smegmatis Soaked With Iron In The Ratio Of 24 Iron Atoms Per Dodecamer
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-08-09 Classification: DNA BINDING PROTEIN Ligands: FE, FE2, MG, CL |
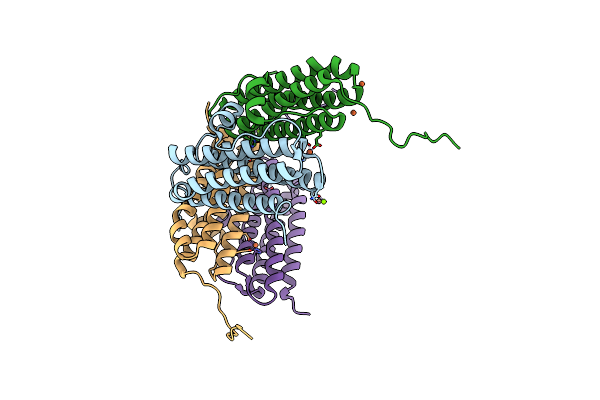 |
Crystal Structure Of The Second Dna-Binding Protein Under Starvation From Mycobacterium Smegmatis Soaked With Iron In The Ratio Of 48 Iron Atoms Per Dodecamer
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-08-09 Classification: DNA BINDING PROTEIN Ligands: FE, MG, CL, FE2 |
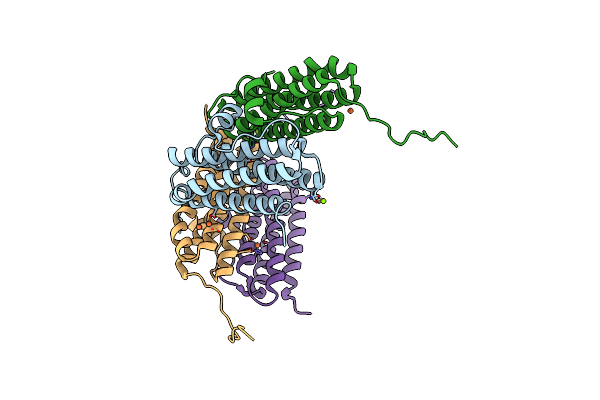 |
Crystal Structure Of The Second Dna-Binding Protein Under Starvation From Mycobacterium Smegmatis Soaked With Iron In The Ratio Of 100 Iron Atoms Per Dodecamer
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2017-08-09 Classification: DNA BINDING PROTEIN Ligands: FE, FE2, MG, CL |
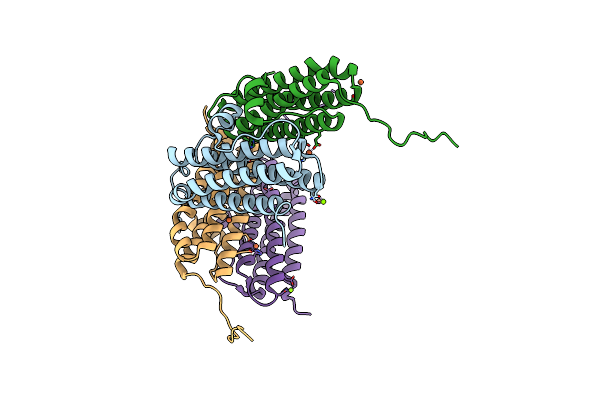 |
Crystal Structure Of The Second Dna-Binding Protein Under Starvation From Mycobacterium Smegmatis Soaked With Iron In The Ratio Of 240 Iron Atoms Per Dodecamer
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2017-08-09 Classification: DNA BINDING PROTEIN Ligands: FE, FE2, MG, CL |

