Search Count: 452
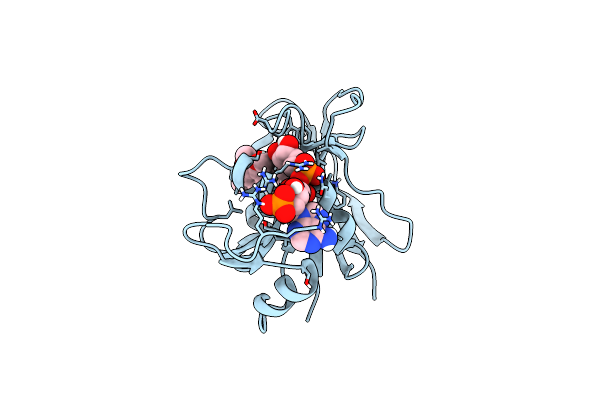 |
Crystal Structure Of Mycobacterium Avium Dihydrofolate Reductase In Complex With Nadph And Trimethoprim
Organism: Mycobacterium avium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NDP, TOP, CL |
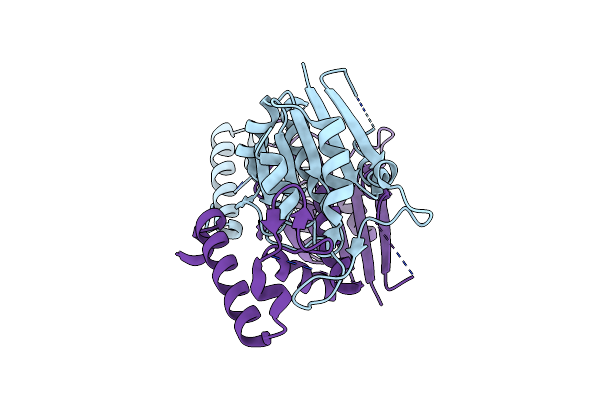 |
Organism: Mycobacterium avium
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-12-13 Classification: TRANSFERASE |
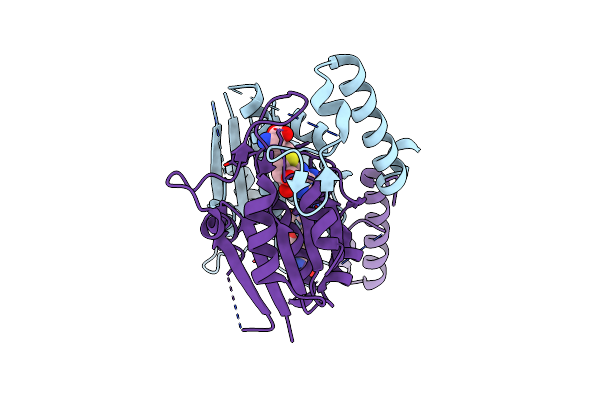 |
Trna (N1G37) Methyltransferase (Trmd) Of Mycobacterium Avium Complexed With S-Adenosyl Homocysteine
Organism: Mycobacterium avium
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-12-13 Classification: TRANSFERASE Ligands: SAH |
 |
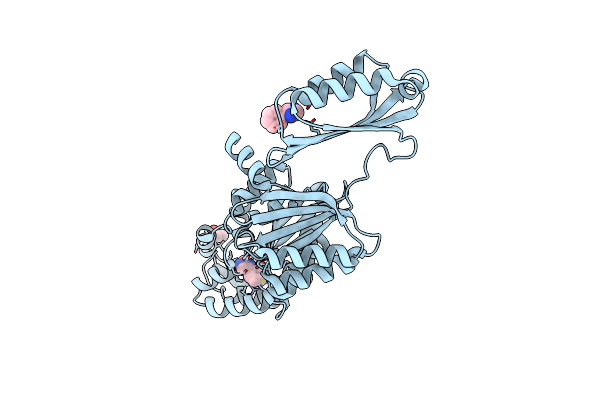
Crystal Structure Of Phosphoserine Phosphatase Serb From Mycobacterium Avium In Complex With 1-(2,4-Dichlorophenyl)-3-Hydroxyurea
Organism: Mycobacterium avium 104
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2022-11-23 Classification: HYDROLASE Ligands: MG, KVU, CL |
 |
Crystal Structure Of Phosphoserine Phosphatase Serb From Mycobacterium Avium In Complex With Phenylimidazole
Organism: Mycobacterium avium 104
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2022-11-23 Classification: HYDROLASE Ligands: MG, PIM, CL, GOL |
 |
Crystal Structure Of A Short Chain Dehydrogenase From Mycobacterium Avium 104
Organism: Mycobacterium avium 104
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-04-13 Classification: OXIDOREDUCTASE |
 |
Organism: Mycobacterium avium (strain 104)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-03-03 Classification: TRANSFERASE Ligands: EDO |
 |
Crystal Structure Of N-Terminus Deletion Mutant Of Mycobacterium Avium Diadenosine 5',5'''-P1,P4-Tetraphosphate Phosphorylase
Organism: Mycobacterium avium
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-08-14 Classification: TRANSFERASE Ligands: SPM, 1PE, PEG |
 |
Crystal Structure Of Enoyl-[Acyl-Carrier-Protein] Reductase From Mycobacterium Avium With Bound Nad
Organism: Mycobacterium avium (strain 104)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-08-14 Classification: OXIDOREDUCTASE Ligands: NAD, ACT, PEG, EDO, PG4 |
 |
Crystal Structure Of Mycobacterium Avium Serb2 In Complex With Serine At Act Domain
Organism: Mycobacterium avium (strain 104)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-05-03 Classification: HYDROLASE Ligands: EDO, PG0, MG, SER |
 |
Crystal Structure Of Mycobacterium Avium Serb2 With Serine Present At Slightly Different Position Near Act Domain
Organism: Mycobacterium avium (strain 104)
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2017-05-03 Classification: HYDROLASE Ligands: MG, EDO, PG0, SER |
 |
Crystal Structure Of Mycobacterium Avium Serb2 In Complex With Serine At Catalytic (Psp) Domain
Organism: Mycobacterium avium (strain 104)
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2017-05-03 Classification: HYDROLASE Ligands: SER, MG, EDO |
 |
Crystal Structure Of Mycobacterium Avium Serb2 Mutant S275A/R279A At Ph 6.6 With Ethylene Glycol Bound At Act- I Domain
Organism: Mycobacterium avium (strain 104)
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2016-11-16 Classification: HYDROLASE Ligands: EDO, MG |
 |
Crystal Structure Of An Adenylyl Cyclase Ma1120-Cat In Complex With Gtp And Calcium From Mycobacterium Avium
Organism: Mycobacterium avium subsp. avium 10-9275
Method: X-RAY DIFFRACTION Release Date: 2016-08-10 Classification: LYASE Ligands: CA, GTP, CL |
 |
Crystal Structure Of Triple Mutant (Kda To Egy) Of Adenylyl Cyclase Ma1120 From Mycobacterium Avium In Complex With Gtp And Calcium Ion
Organism: Mycobacterium avium subsp. avium 10-9275
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-08-10 Classification: LYASE Ligands: GTP, CA |
 |
Crystal Structure Of Triple Mutant (Kda To Egy) Of An Adenylyl Cyclase Ma1120 From Mycobacterium Avium In Complex With Atp And Calcium Ion
Organism: Mycobacterium avium subsp. avium 10-9275
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-08-10 Classification: LYASE Ligands: ATP, CA |
 |
Crystal Structure Of An Adenylyl Cyclase Ma1120 From Mycobacterium Avium In Complex With Atp And Calcium Ion
Organism: Mycobacterium avium subsp. avium 10-9275
Method: X-RAY DIFFRACTION Release Date: 2016-08-10 Classification: LYASE Ligands: ATP, CA, EDO |
 |
Organism: Mycobacterium avium (strain 104)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: MG, EDO |
 |
Organism: Mycobacterium avium (strain 104)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2016-04-27 Classification: HYDROLASE Ligands: MG |
 |
Organism: Mycobacterium avium (strain 104)
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2016-04-27 Classification: HYDROLASE Ligands: MG |

