Search Count: 22
 |
Crystal Structure Of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-02-21 Classification: OXIDOREDUCTASE Ligands: ZN, EDO, PEG, PGE, PG4 |
 |
Crystal Structure Of Alcohol Dehydrogenase From Arabidopsis Thaliana In Complex With Nadh
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-02-21 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, SO4, PG4, PEG, EDO |
 |
1.93 A Resolution X-Ray Crystal Structure Of The Transcriptional Regulator Srnr From Streptomyces Griseus
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-05-25 Classification: DNA BINDING PROTEIN Ligands: ACT, NA |
 |
Crystal Structure Of Malus Domestica Double Bond Reductase (Mddbr) Apo Form
Organism: Pyrus ussuriensis x pyrus communis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-02-03 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |
 |
Crystal Structure Of Malus Domestica Double Bond Reductase (Mddbr) In Complex With Nadph
Organism: Malus domestica
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-02-03 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP, MPD, EDO, PGR, NA |
 |
Crystal Structure Of Malus Domestica Double Bond Reductase (Mddbr) Ternary Complex
Organism: Malus domestica
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2021-02-03 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP, MPD, EDO, FER, NA |
 |
Crystal Structure Of Myricetin Covalently Bound To The Main Protease (3Clpro/Mpro) Of Sars-Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2021-01-13 Classification: VIRAL PROTEIN Ligands: EDO, CL, MYC, NA |
 |
1.50 A Resolution 3-Methylcatechol (3-Methylbenzene-1,2-Diol) Inhibited Sporosarcina Pasteurii Urease
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: EDO, SO4, NI, OH |
 |
1.89 A Resolution 4-Methylcatechol (4-Methylbenzene-1,2-Diol) Inhibited Sporosarcina Pasteurii Urease
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: EDO, SO4, NI, OH |
 |
2.23 A Resolution 3,4-Dimethylcatechol (3,4-Dimethylbenzene-1,2-Diol) Inhibited Sporosarcina Pasteurii Urease
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: EDO, SO4, NI, OH |
 |
1.61 A Resolution 3,5-Dimethylcatechol (3,5-Dimethylbenzene-1,2-Diol) Inhibited Sporosarcina Pasteurii Urease
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: EDO, SO4, NI, OH |
 |
1.65 A Resolution 4,5-Dimethylcatechol (4,5-Dimethylbenzene-1,2-Diol) Inhibited Sporosarcina Pasteurii Urease
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: EDO, SO4, NI, OH |
 |
1.55 A Resolution 3,6-Dimethylcatechol (3,6-Dimethylbenzene-1,2-Diol) Inhibited Sporosarcina Pasteurii Urease
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: EDO, SO4, NI, OH |
 |
Solution Structure Of The Ni Metallochaperone Hypa From Helicobacter Pylori
Organism: Helicobacter pylori j99
Method: SOLUTION NMR Release Date: 2018-10-10 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
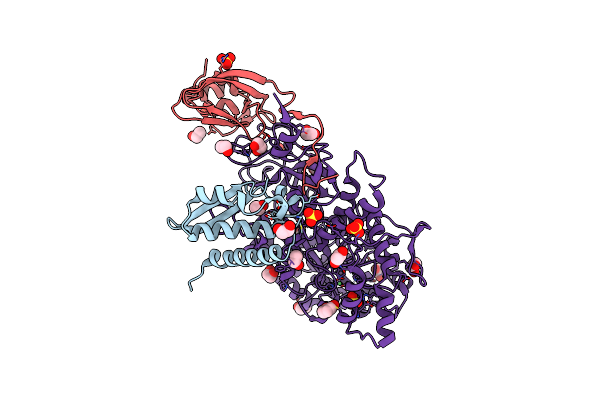
1.28 A Resolution Of Sporosarcina Pasteurii Urease Inhibited In The Presence Of Nbpt
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2018-03-07 Classification: HYDROLASE Ligands: EDO, SO4, NI, 9XN |
 |
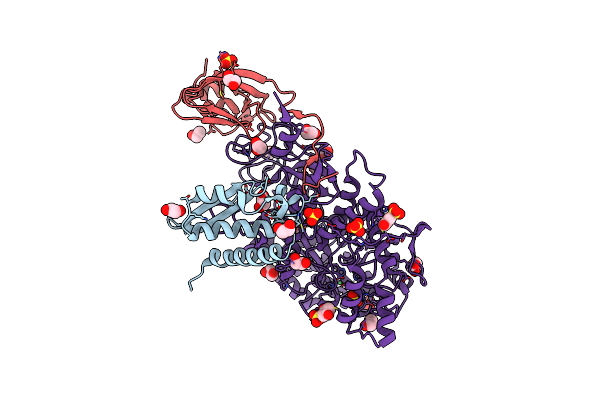
1.50 A Resolution Catechol (1,2-Dihydroxybenzene) Inhibited Sporosarcina Pasteurii Urease
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-12-07 Classification: HYDROLASE Ligands: SO4, EDO, NI, CAQ, OH |
 |
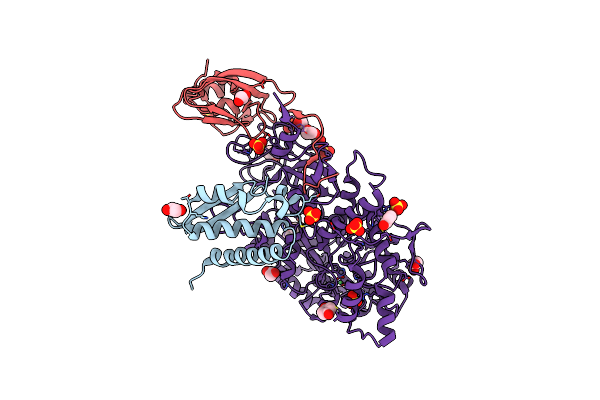
1.75 A Resolution 2,5-Dihydroxybenzensulfonate Inhibited Sporosarcina Pasteurii Urease
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-03-23 Classification: HYDROLASE Ligands: EDO, SO4, NI, OH, DBX |
 |
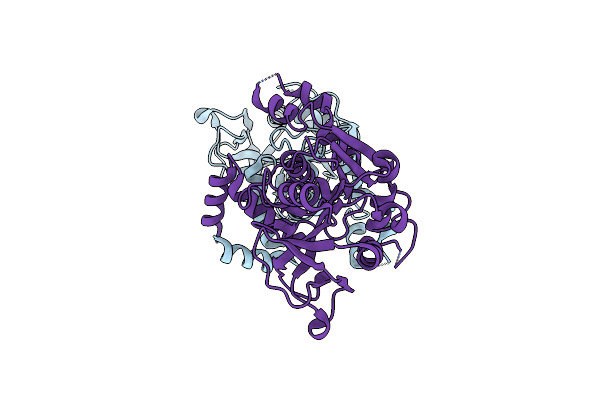
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2016-03-23 Classification: HYDROLASE Ligands: EDO, SO4, NI, OH, HQE |
 |
Crystal Structure Of Glucose-1-Phosphate Uridylyltransferase Galu From Erwinia Amylovora.
Organism: Erwinia amylovora
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2016-01-20 Classification: TRANSFERASE |
 |
Organism: Erwinia amylovora
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2016-01-20 Classification: HYDROLASE Ligands: SO4 |

