Search Count: 27
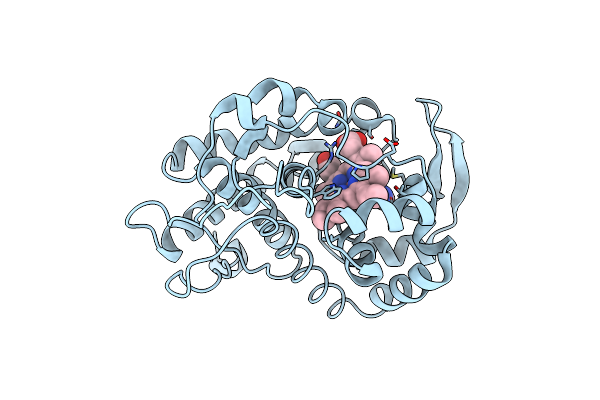 |
Crystal Structure Of Cytochrome C Peroxidase With A Proposed Electron Transfer Pathway Excised To Form A Ligand Binding Channel.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2002-03-06 Classification: OXIDOREDUCTASE Ligands: HEM, BZI |
 |
Crystal Structure Of Cytochrome C Peroxidase With A Proposed Electron Transfer Pathway Excised To Form A Ligand Binding Channel.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2002-03-06 Classification: OXIDOREDUCTASE Ligands: HEM |
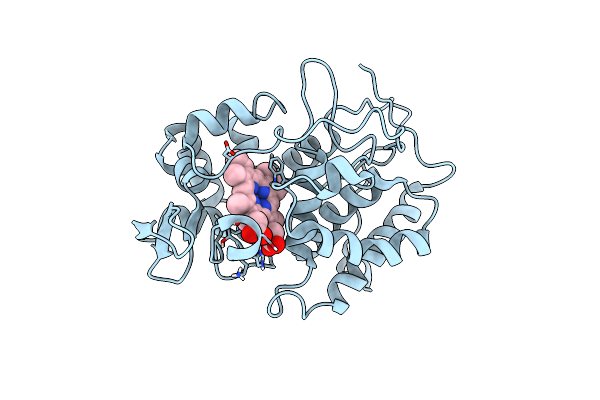 |
Probing The Strength And Character Of An Asp-His-X Hydrogen Bond By Introducing Buried Charges
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1998-11-25 Classification: OXIDOREDUCTASE Ligands: HEM |
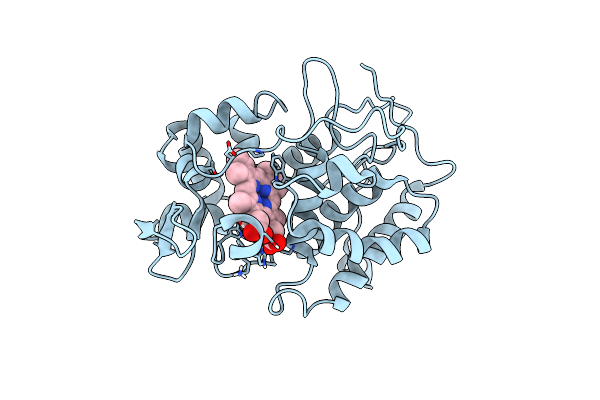 |
Probing The Strength And Character Of An Asp-His-X Hydrogen Bond By Introducing Buried Charges
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1998-11-25 Classification: OXIDOREDUCTASE Ligands: HEM |
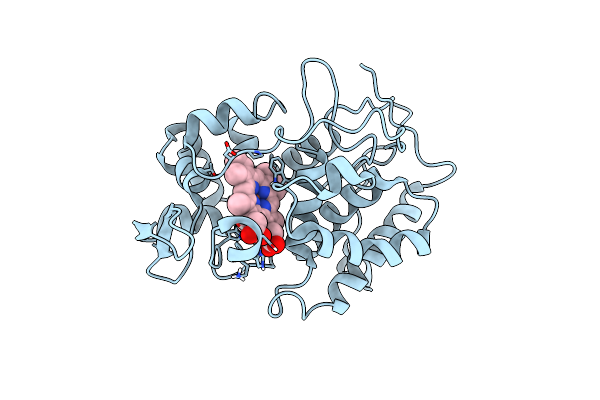 |
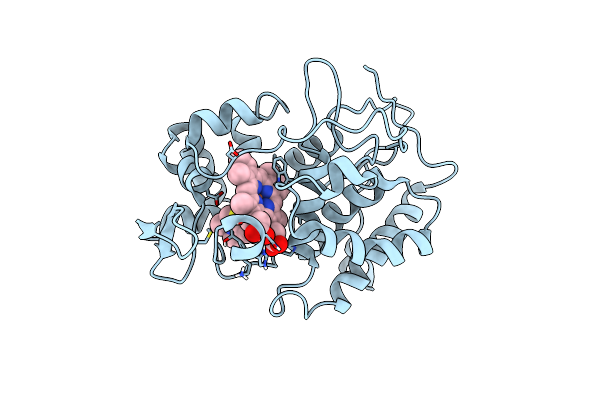
Probing The Strength And Character Of An Asp-His-X Hydrogen Bond By Introducing Buried Charges
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1998-11-25 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Variation In The Strength Of A Ch To O Hydrogen Bond In An Artificial Protein Cavity (2,3,4-Trimethyl-1,3-Thiazole)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-09-04 Classification: OXIDOREDUCTASE Ligands: HEM, TMT |
 |
Variation In The Strength Of A Ch To O Hydrogen Bond In An Artificial Protein Cavity (3,4,5-Trimethylthiazole)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-09-04 Classification: OXIDOREDUCTASE Ligands: HEM, TMZ |
 |
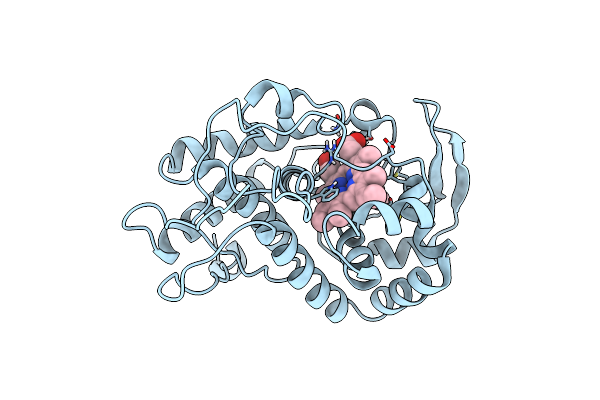
Specificity Of Ligand Binding To A Buried Polar Cavity At The Active Site Of Cytochrome C Peroxidase (3-Methylthiazole)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-09-04 Classification: OXIDOREDUCTASE Ligands: HEM, 3MT |
 |
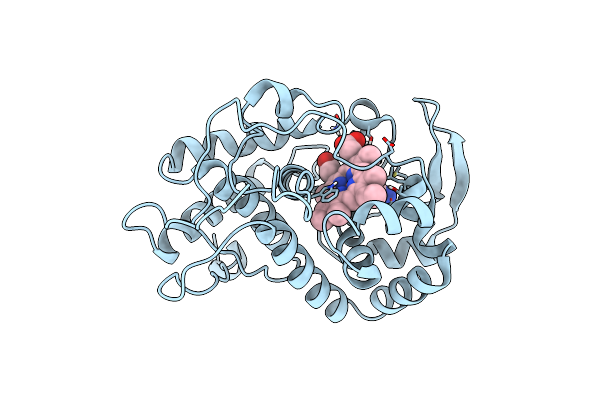
Specificity Of Ligand Binding To A Buried Polar Cavity At The Active Site Of Cytochrome C Peroxidase (3,4-Dimethylthiazole)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-09-04 Classification: OXIDOREDUCTASE Ligands: HEM, DTI |
 |
Specificity Of Ligand Binding To A Buried Polar Cavity At The Active Site Of Cytochrome C Peroxidase (Aniline)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-09-04 Classification: OXIDOREDUCTASE Ligands: HEM, ANL |
 |
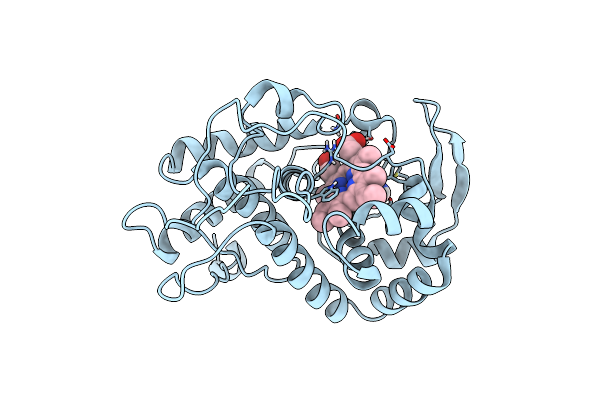
Specificity Of Ligand Binding To A Buried Polar Cavity At The Active Site Of Cytochrome C Peroxidase (3-Aminopyridine)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-09-04 Classification: OXIDOREDUCTASE Ligands: HEM, 3AP |
 |
Specificity Of Ligand Binding To A Buried Polar Cavity At The Active Site Of Cytochrome C Peroxidase (4-Aminopyridine)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-09-04 Classification: OXIDOREDUCTASE Ligands: HEM, 4AP |
 |
Specificity Of Ligand Binding To A Buried Polar Cavity At The Active Site Of Cytochrome C Peroxidase (2-Amino-4-Methylthiazole)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-09-04 Classification: OXIDOREDUCTASE Ligands: HEM, 24T |
 |
Specificity Of Ligand Binding To A Buried Polar Cavity At The Active Site Of Cytochrome C Peroxidase (1-Vinylimidazole)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-09-04 Classification: OXIDOREDUCTASE Ligands: HEM, NVI |
 |
Specificity Of Ligand Binding To A Buried Polar Cavity At The Active Site Of Cytochrome C Peroxidase (Indoline)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-09-04 Classification: OXIDOREDUCTASE Ligands: HEM, IDM |
 |
Specificity Of Ligand Binding To A Buried Polar Cavity At The Active Site Of Cytochrome C Peroxidase (Imidazo[1,2-A]Pyridine)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-09-04 Classification: OXIDOREDUCTASE Ligands: HEM, MPI |
 |
Specificity Of Ligand Binding To A Buried Polar Cavity At The Active Site Of Cytochrome C Peroxidase (2-Amino-5-Methylthiazole)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-09-04 Classification: OXIDOREDUCTASE Ligands: HEM, 25T |
 |
Specificity Of Ligand Binding To A Buried Polar Cavity At The Active Site Of Cytochrome C Peroxidase (2-Aminopyridine)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-09-04 Classification: OXIDOREDUCTASE Ligands: HEM, 2AP |
 |
Variation In The Strength Of A Ch To O Hydrogen Bond In An Artificial Protein Cavity (2-Ethylimidazole)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-09-04 Classification: OXIDOREDUCTASE Ligands: HEM, 2EZ |
 |
Specificity Of Ligand Binding To A Buried Polar Cavity At The Active Site Of Cytochrome C Peroxidase (Imidazole)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-09-04 Classification: OXIDOREDUCTASE Ligands: HEM, IMD |

