Search Count: 20
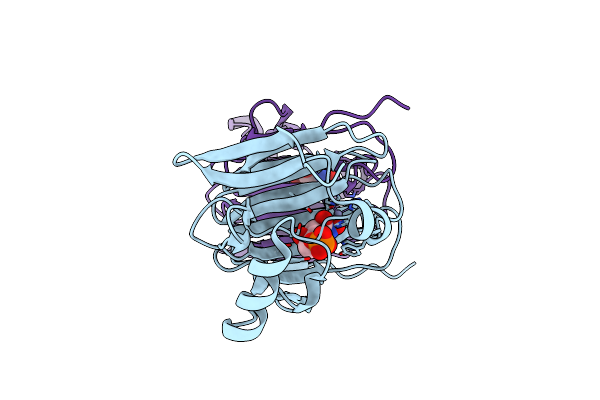 |
The Crystal Structure Of Phosphoglucose Isomerase From Pyrococcus Furiosus In Complex With 5-Phospho-D-Arabinonohydroxamate And Zinc
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-04-11 Classification: ISOMERASE Ligands: ZN, PAN |
 |
The Crystal Structure Of Phosphoglucose Isomerase From Pyrococcus Furiosus In Complex With Sorbitol 6-Phosphate And Zinc
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2006-04-11 Classification: ISOMERASE Ligands: S6P, ZN |
 |
The Crystal Structure Of Phosphoglucose Isomerase From Pyrococcus Furiosus In Complex With Fructose 6-Phosphate And Zinc
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-04-11 Classification: ISOMERASE Ligands: ZN, F6R |
 |
The Crystal Structure Of Phosphoglucose Isomerase From Pyrococcus Furiosus In Complex With Mannose 6-Phosphate And Zinc
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-04-11 Classification: ISOMERASE Ligands: ZN, M6P |
 |
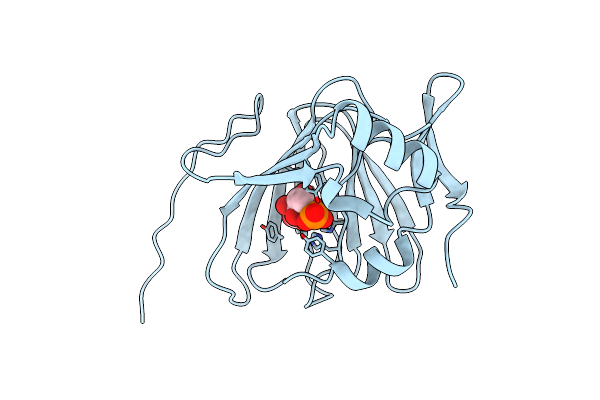
The Crystal Structure Of Pyrococcus Furiosus Phosphoglucose Isomerase With Bound 5-Phospho-D-Arabinonate And Manganese
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2004-10-12 Classification: ISOMERASE Ligands: MN, PA5 |
 |
Crystal Structure Of Phosphoglucose Isomerase From Pyrococcus Furiosus With Bound 5-Phospho-D-Arabinonate
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2004-10-12 Classification: METAL BINDING PROTEIN Ligands: PA5 |
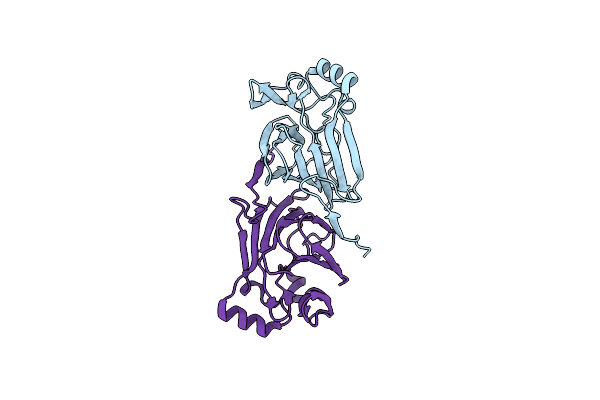 |
Crystal Structure Of Pyrococcus Furiosus Phosphoglucose Isomerase Free Enzyme
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2004-10-12 Classification: ISOMERASE |
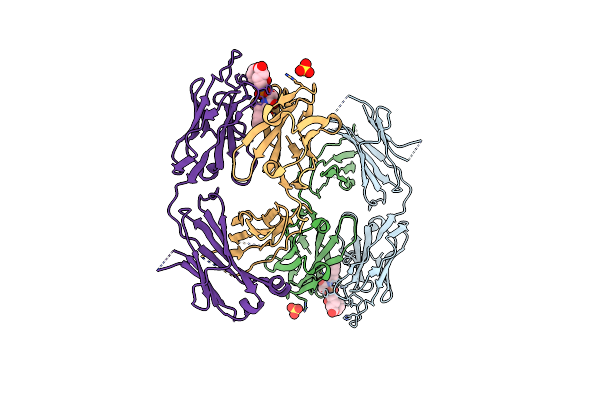 |
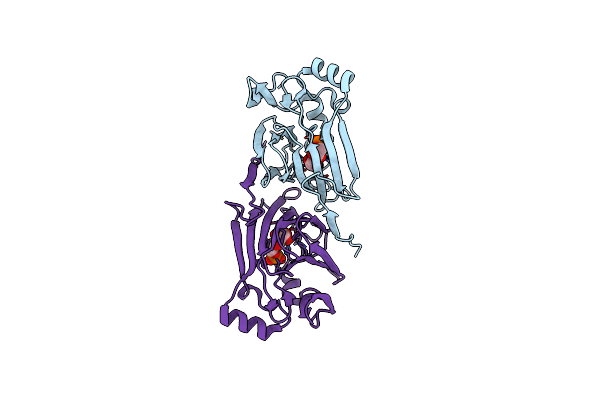
The Structure Of The Complex Of The Fab Fragment Of The Esterolytic Antibody Ms6-164 And A Transition-State Analog
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2003-09-23 Classification: IMMUNE SYSTEM Ligands: SO4, HAL |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2003-09-23 Classification: IMMUNE SYSTEM |
 |
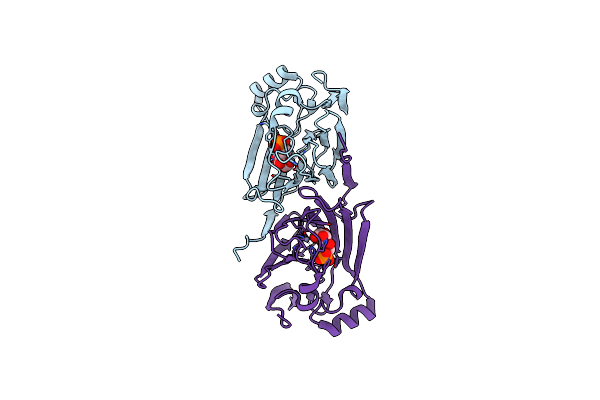
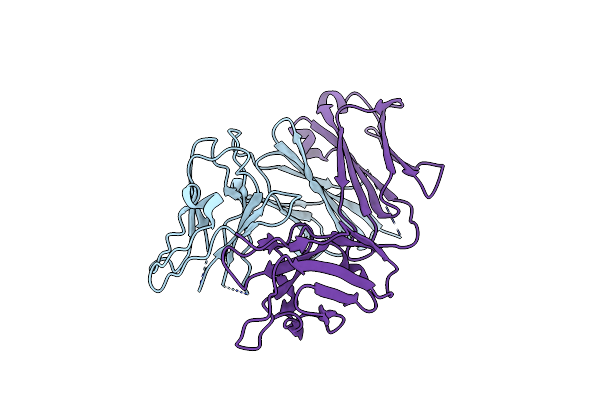
Crystal Structure Of The Complex Of The Fab Fragment Of Esterolytic Antibody Ms5-393 And A Transition-State Analog
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2003-09-23 Classification: IMMUNE SYSTEM Ligands: HAL |
 |
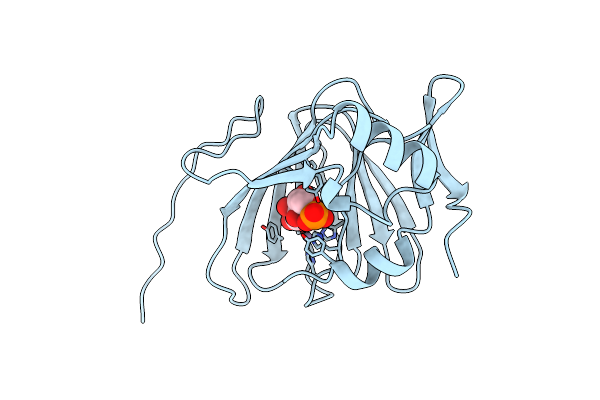
High Resolution Crystal Structure Of The Fab Fragment Of The Esterolytic Antibody Ms6-126
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2003-09-23 Classification: IMMUNE SYSTEM Ligands: PO4, GOL |
 |
High Resolution Crystal Structure Of The Complex Of The Fab Fragment Of Esterolytic Antibody Ms6-12 And A Transition-State Analog
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2003-09-23 Classification: IMMUNE SYSTEM Ligands: SO4, HAL |
 |
1.22 Angstrom Resolution Crystal Structure Of The Fab Fragment Of Esterolytic Antibody Ms6-12
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2003-09-23 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-08-26 Classification: TRANSFERASE |
 |
Crystal Structure Of Streptogramin A Acetyltransferase With Acetyl-Coa Bound
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2003-08-26 Classification: TRANSFERASE Ligands: ACO |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-08-26 Classification: TRANSFERASE Ligands: DOL |
 |
Quadruple Mutant Q92C, N146F, Y168F, I172V Type Iii Cat Complexed With Fusidic Acid. Crystals Grown At Ph 6.3. X-Ray Data Collected At Room Temperature
Organism: Shigella flexneri
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1995-10-15 Classification: TRANSFERASE (ACYLTRANSFERASE) Ligands: CO, FUA |
 |
Replacement Of Catalytic Histidine-195 Of Chloramphenicol Acetyltransferase: Evidence For A General Base Role For Glutamate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1994-01-31 Classification: TRANSFERASE(ACYLTRANSFERASE) Ligands: CO, BME |
 |
Alternative Binding Modes For Chloramphenicol And 1-Substituted Chloramphenicol Analogues Revealed By Site-Directed Mutagenesis And X-Ray Crystallography Of Chloramphenicol Acetyltransferase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1992-01-15 Classification: TRANSFERASE (ACYLTRANSFERASE) Ligands: CO, CLM |
 |
Evidence For Transition-State Stabilization By Serine-148 In The Catalytic Mechanism Of Chloramphenicol Acetyltransferase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 1990-07-15 Classification: TRANSFERASE (ACYLTRANSFERASE) Ligands: CO, CLM |

