Search Count: 25
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: PROTEIN FIBRIL |
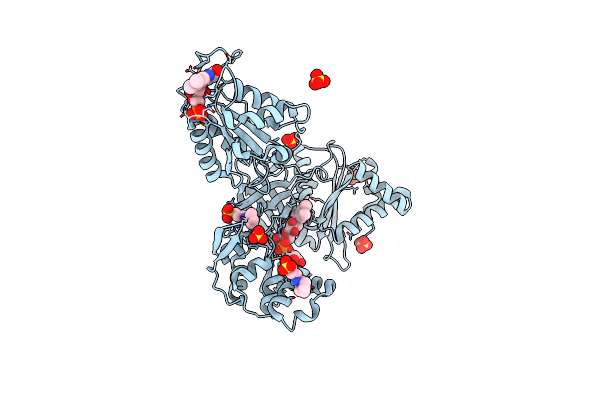 |
The Nadph-Dependent Sulfite Reductase Flavoprotein Adopts An Extended Conformation That Is Unique To This Diflavin Reductase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2019-02-27 Classification: FLAVOPROTEIN Ligands: FAD, FMN, SO4, CXS |
 |
Organism: Homo sapiens
Method: SOLID-STATE NMR Release Date: 2017-09-27 Classification: PROTEIN FIBRIL |
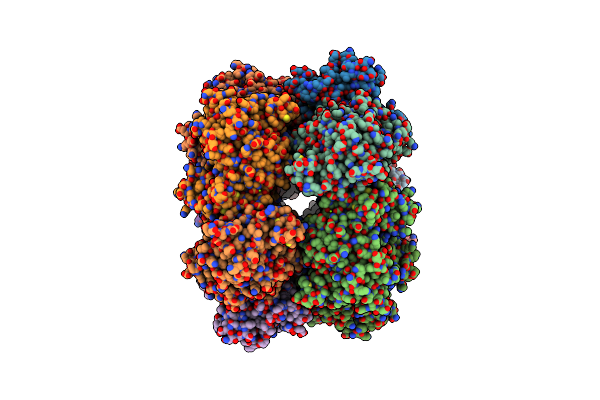 |
B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes And Basis For Isoenzyme Specific Regulation: Structure Of Gs-Q
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2013-11-13 Classification: LIGASE Ligands: MG, GLN, PO4 |
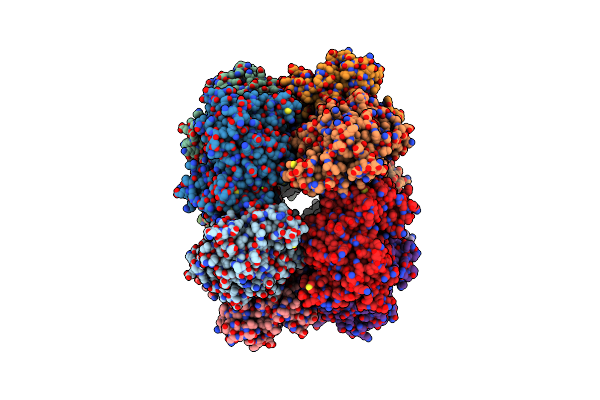 |
B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes And Basis For Isoenzyme Specific Regulation: Structure Of Apo Form Of Gs
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-11-13 Classification: LIGASE Ligands: MG, SO4 |
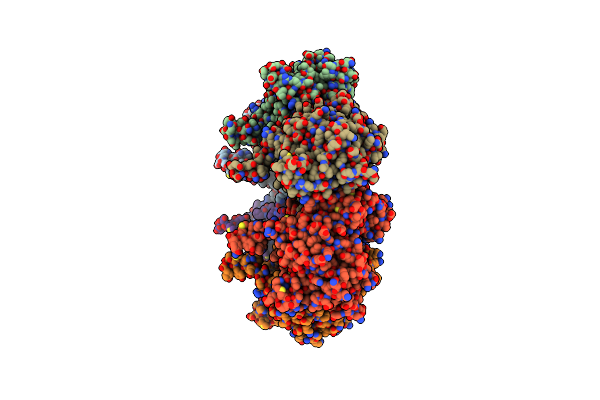 |
B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes And Basis For Isoenzyme Specific Regulation: Form Two Of Gs-1
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2013-11-13 Classification: LIGASE Ligands: MG, GLN |
 |
B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes And Basis For Isoenzyme Specific Regulation: Structure Of The Transition State Complex
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2013-11-06 Classification: LIGASE Ligands: P3S, ADP, MG |
 |
B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes And Basis For Isoenzyme Specific Regulation: Structure Of Gs-Glutamate-Amppcp Complex
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2013-10-30 Classification: LIGASE Ligands: GLU, ADP, MG, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-07-31 Classification: TRANSPORT PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2007-07-31 Classification: REGULATOR Ligands: DTT |
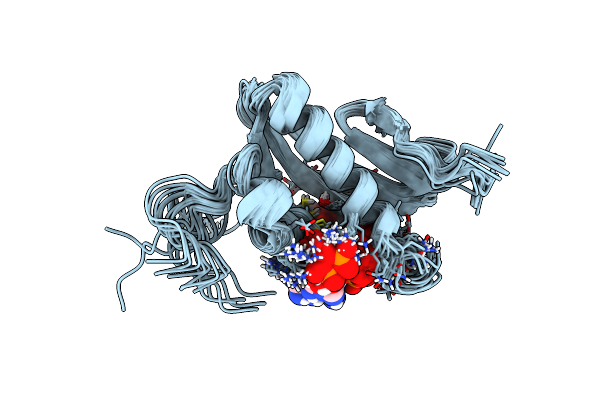 |
Nmr Solution Structure Of A Gcn5-Like Putative N-Acetyltransferase From Staphylococcus Aureus Complexed With Acetyl-Coa. Northeast Structural Genomics Consortium Target Zr31
Organism: Staphylococcus aureus subsp. aureus
Method: SOLUTION NMR Release Date: 2006-11-28 Classification: TRANSFERASE Ligands: ACO |
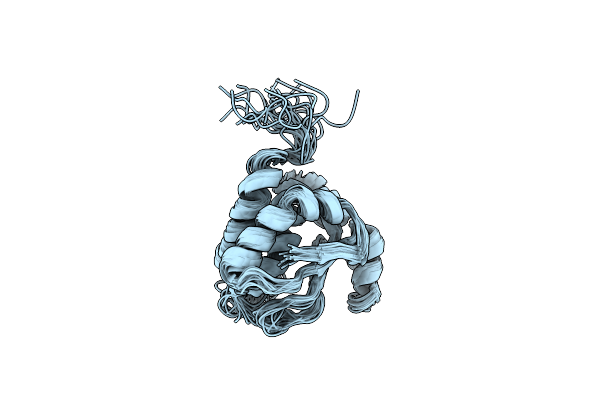 |
Solution Structure Of Tm1509 From Thermotoga Maritima: Vt1, A Nesgc Target Protein
Organism: Thermotoga maritima
Method: SOLUTION NMR Release Date: 2005-01-04 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
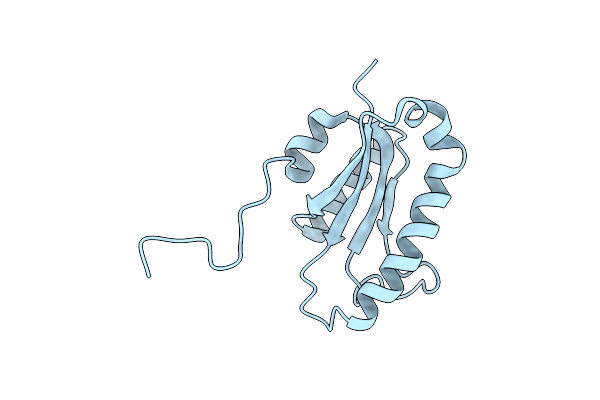 |
The Solution Structure Of The Archaeglobus Fulgidis Protein Af2095. Northeast Structural Genomics Consortium Target Gr4
Organism: Archaeoglobus fulgidus
Method: SOLUTION NMR Release Date: 2004-11-16 Classification: Structural Genomics, Unknown function |
 |
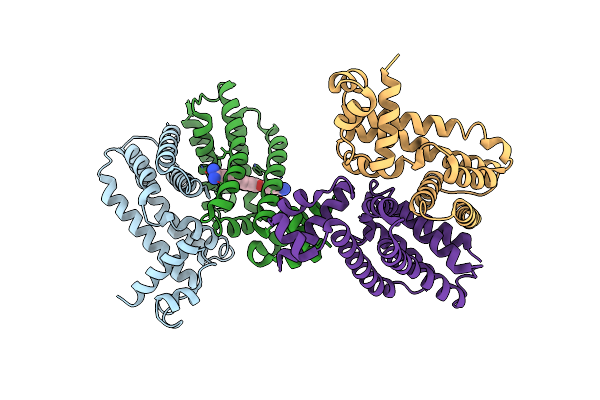
Crystal Structure Of The Multidrug Binding Transcriptional Repressor Qacr Bound To Pentamadine
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2004-06-01 Classification: TRANSCRIPTION Ligands: SO4, PNT |
 |
Crystal Structure Of The Multidrug Binding Protein Qacr Bound To The Diamidine Hexamidine
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2004-06-01 Classification: TRANSCRIPTION Ligands: SO4, DID |
 |
Nmr Solution Structure Of A Gcn5-Like Putative N-Acetyltransferase From Staphylococcus Aureus. Northeast Structural Genomics Consortium Target Zr31
Organism: Staphylococcus aureus
Method: SOLUTION NMR Release Date: 2004-03-09 Classification: TRANSFERASE |
 |
Solution Structure Of 30S Ribosomal Protein S27E From Archaeoglobus Fulgidus: Gr2, A Nesg Target Protein
Organism: Archaeoglobus fulgidus
Method: SOLUTION NMR Release Date: 2003-09-16 Classification: RIBOSOME |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2003-06-03 Classification: STRUCTURAL GENOMICS, Unknown function |
 |
The Structure Of Cyclin-Dependent Kinase 2 (Cdk2) In Complex With An Oxindole Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2001-01-17 Classification: TRANSFERASE, CELL CYCLE Ligands: 106 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2001-01-17 Classification: TRANSFERASE, CELL CYCLE Ligands: 107 |

