Search Count: 54
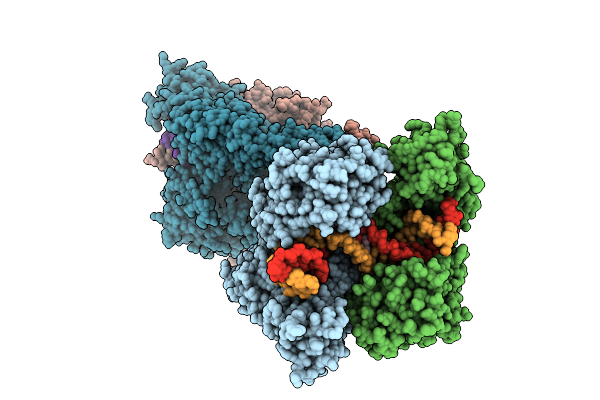 |
Crystal Structure Of Topoisomerase Iv From Klebsiella Pneumoniae In Complex With Dna And Bwc0977, A Dual-Targeting Broad-Spectrum Novel Bacterial Topoisomerase Inhibitor.
Organism: Klebsiella pneumoniae subsp. pneumoniae mgh 78578, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: ISOMERASE/DNA/ISOMERASE INHIBITOR Ligands: A1L5V |
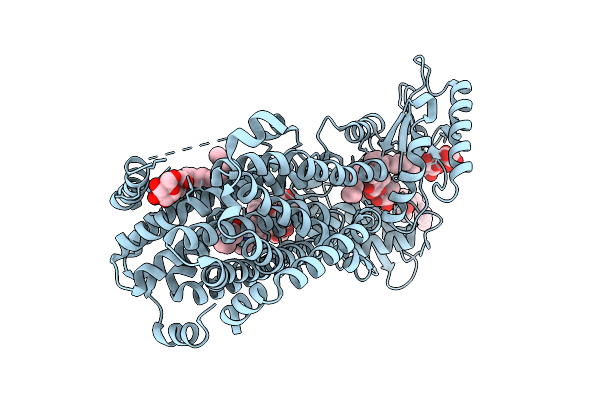 |
Crystal Structure Of Mycolic Acid Transporter Mmpl3 From Mycobacterium Smegmatis Complexed With Indolcarboxamide
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: A1L5O, LMT |
 |
Crystal Structure Of Aminoglycoside Efflux Transporter Mexy From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: SO4 |
 |
Crystal Structure Of Multidrug Efflux Transporter Oqxb From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: MEMBRANE PROTEIN Ligands: LMT, GOL |
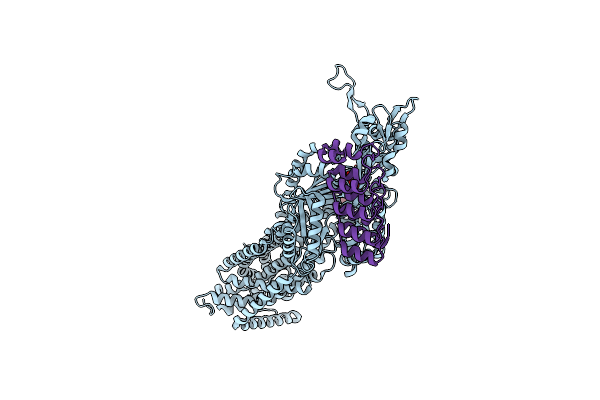 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN Ligands: MIY |
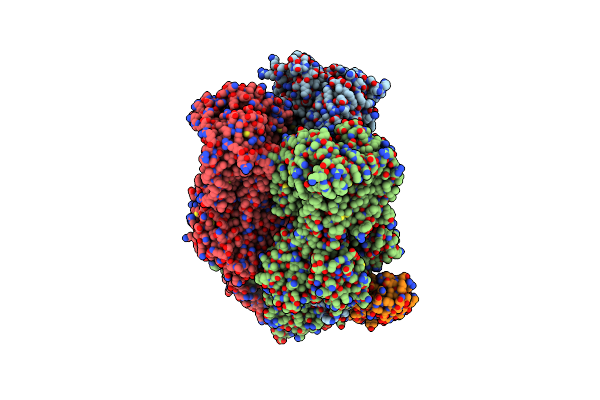 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
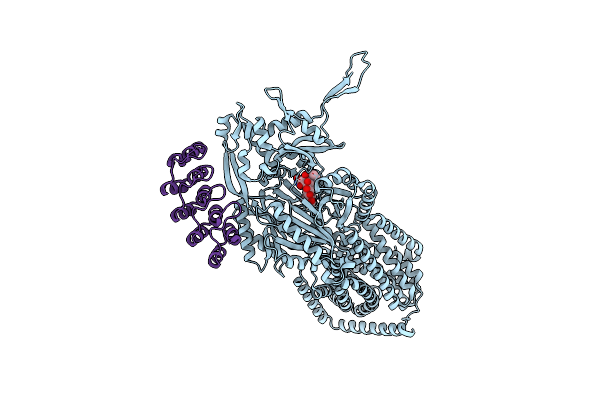
Single Particle Cryo-Em Structure Of The Multidrug Efflux Pump Oqxb From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
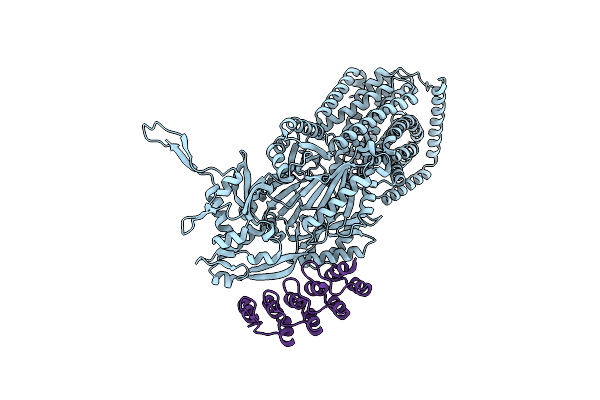
Organism: Synthetic construct, Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN Ligands: MIY |
 |
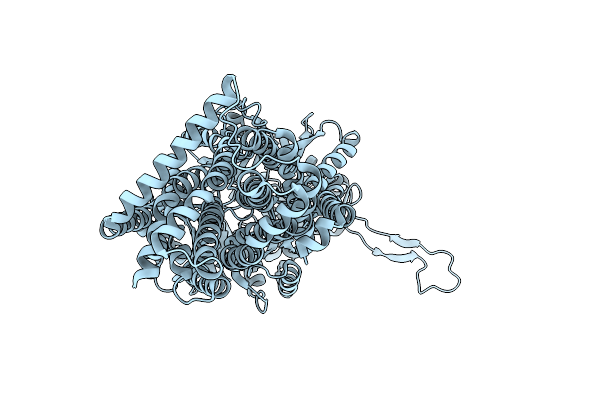
Organism: Synthetic construct, Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-10-09 Classification: TRANSFERASE Ligands: PLS, A1L3N |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2024-10-09 Classification: TRANSFERASE Ligands: PGE |
 |
Crystal Structure Of The Multidrug Efflux Transporter Bpeb From Burkholderia Pseudomallei
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2023-07-19 Classification: MEMBRANE PROTEIN Ligands: UMQ, PG4 |
 |
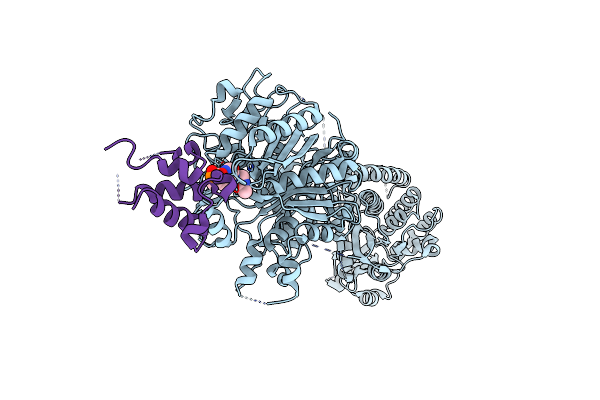
Crystal Structure Of The Multidrug Effulx Transporter Bpef From Burkholderia Pseudomallei.
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-07-19 Classification: MEMBRANE PROTEIN Ligands: LMT |
 |
The Structure Of The Gfsa Ksq-At Didomain In Complex With The Gfsa Acp Domain
Organism: Streptomyces graminofaciens
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2023-05-31 Classification: LYASE Ligands: 9EF |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-12-07 Classification: RNA BINDING PROTEIN Ligands: 9JT |
 |
Crystal Structure Of Ythdf1 Yth Domain In Complex With The Ebsulfur Derivative Compound 7
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2022-11-23 Classification: RNA BINDING PROTEIN Ligands: JVF, EDO, SCN |

