Search Count: 232
 |
Active Conformation Of A Redox-Regulated Glycoside Hydrolase (Capgh2B) From The Gh2 Family
Organism: Metagenome
Method: ELECTRON MICROSCOPY Resolution:2.62 Å Release Date: 2025-11-12 Classification: HYDROLASE |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group I213 At 2.05 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, GOL |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E553Q Mutant) From The Gh2 Family In The Space Group I213 At 2.6 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4 |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group I213 At 2.75 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, TAU |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group R3 At 2.45 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2025-11-12 Classification: HYDROLASE |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group I212121 At 2.65 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, PEG, PG4 |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E553Q Mutant) From The Gh2 Family In The Space Group P3121 At 3.05 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, ACT, MLI |
 |
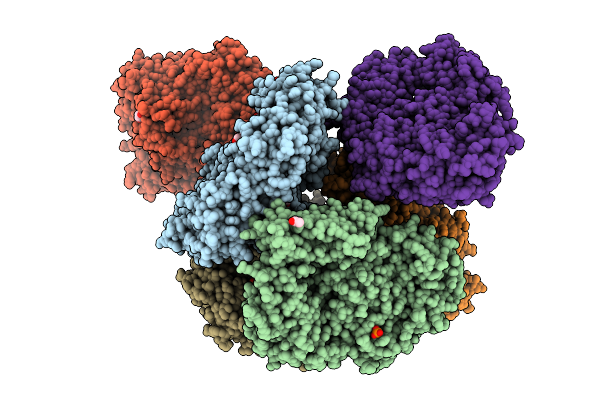
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group P1 At 2.40 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, GOL |
 |
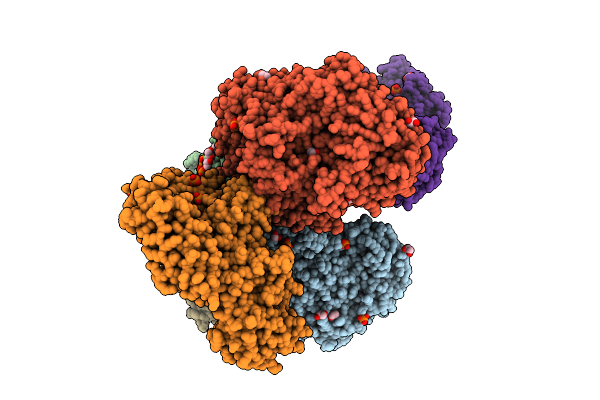
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group P1 At 2.15 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, GOL, ACT |
 |
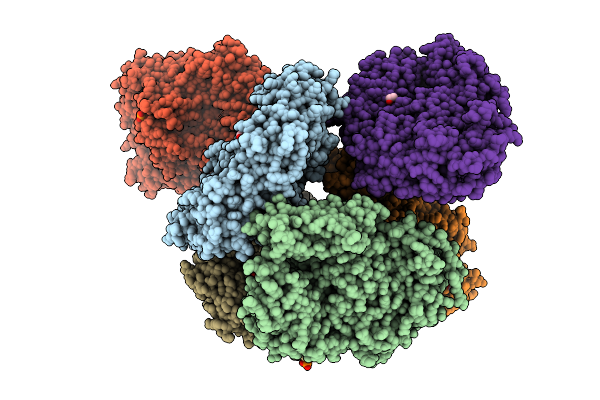
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E553Q Mutant) From The Gh2 Family In The Space Group P1 At 2.25 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, EDO |
 |
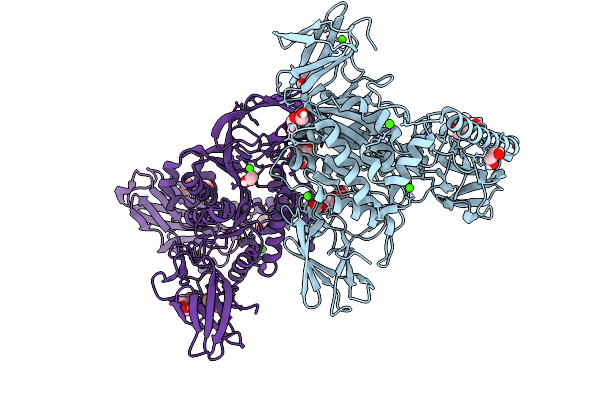
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E465A Mutant) From The Gh2 Family In The Space Group P1 At 3.1 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, EDO |
 |
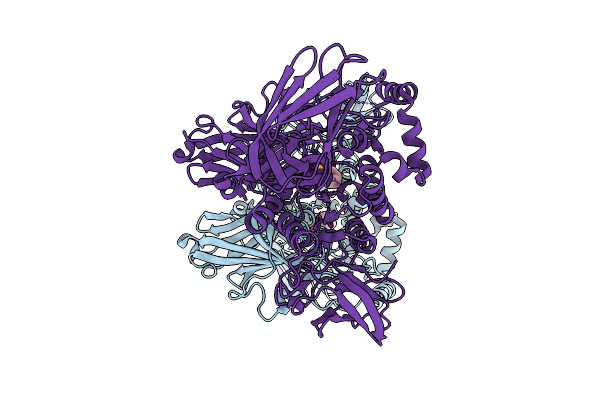
Deacetylase Fi8 Utilizes Unconventional Variant Of A Catalytic Triad: The Diluted Triad
Organism: Flavimarina sp. hel_i_48
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2025-08-27 Classification: HYDROLASE Ligands: PGE, PEG, EDO, CA |
 |
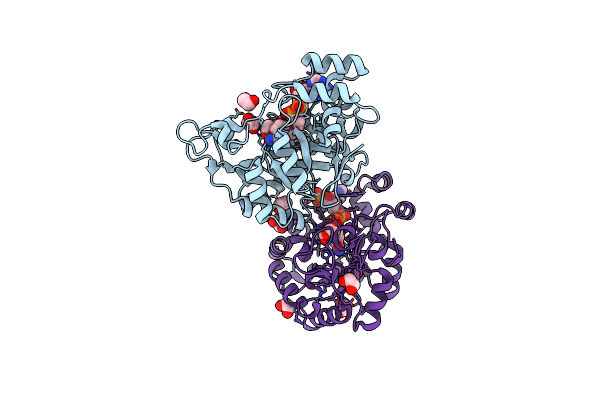
Crystal Structure Of A Ce20 Carbohydrate Acetylesterase From Pedobacter Psychrotolerans (Ppce20_Ii), With Ancillary Domain
Organism: Pedobacter psychrotolerans
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-08-13 Classification: HYDROLASE Ligands: PGE, MG, PMS |
 |
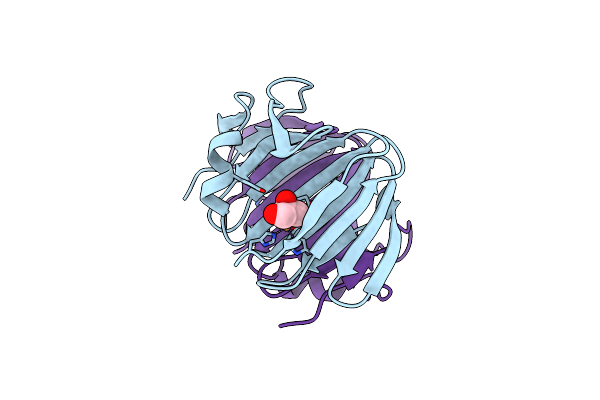
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: NAP, EDO |
 |
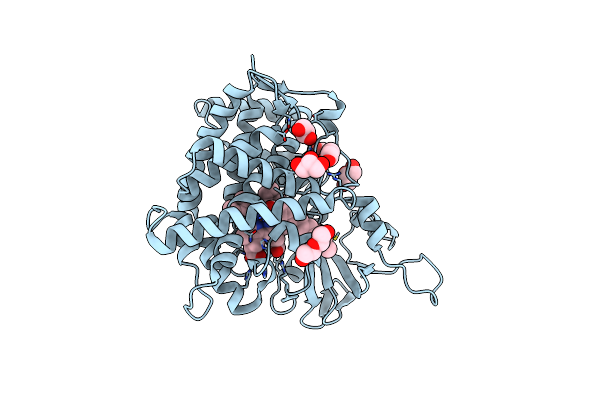
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-12-11 Classification: OXIDOREDUCTASE Ligands: GOL, CU |
 |
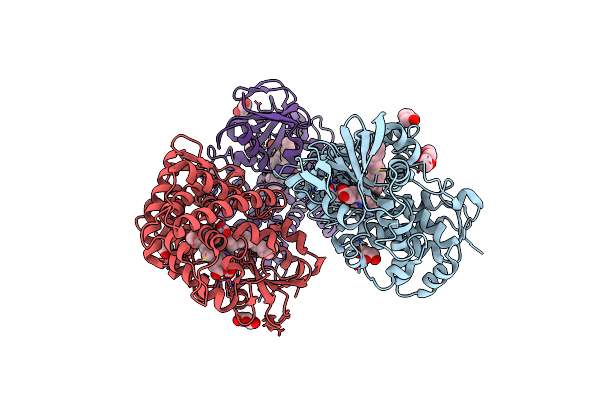
Crystal Structure Of A Fatty Acid Decarboxylase From Kocuria Marina In Complex With Myristic Acid
Organism: Kocuria marina
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: MYR, HEM, GOL, PEG |
 |
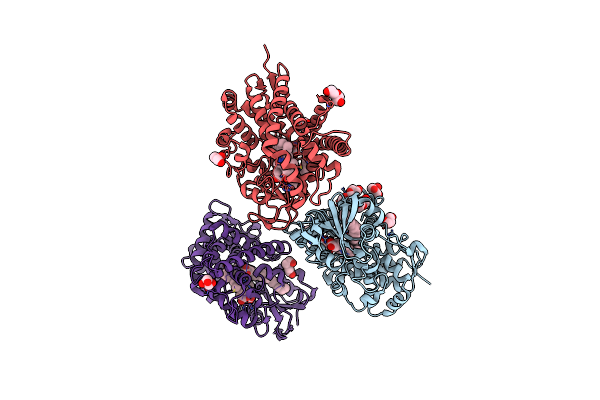
Crystal Structure Of A Fatty Acid Decarboxylase From Corynebacterium Lipophiloflavum In Complex With Palmitic Acid
Organism: Corynebacterium lipophiloflavum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: CL, HEM, PLM, PEG, GOL, PGE |
 |
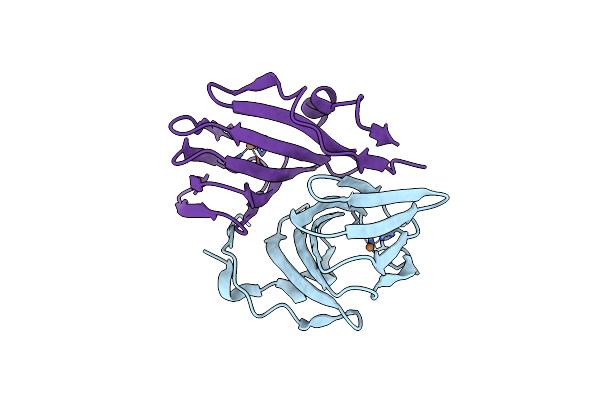
Crystal Structure Of A Fatty Acid Decarboxylase From Corynebacterium Lipophiloflavum In Complex With Oleic Acid
Organism: Corynebacterium lipophiloflavum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: HEM, OLA, PEG, GOL, CL, PGE |
 |
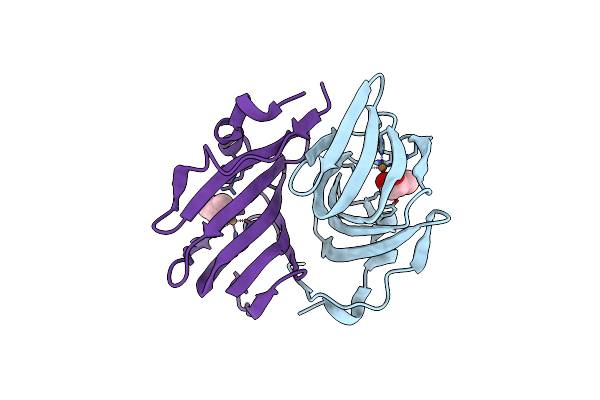
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: GOL, CU |

