Search Count: 198
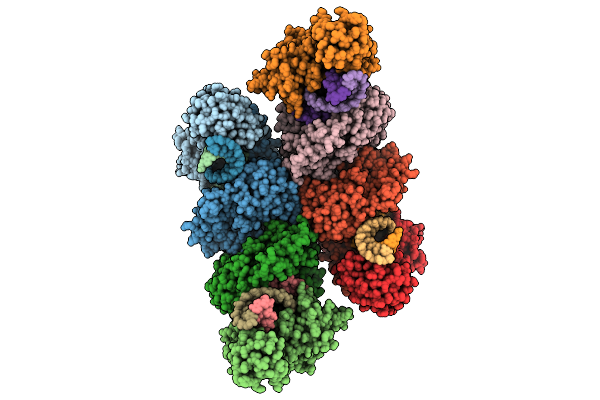 |
Cryo-Em Structure Of Filament Form Acidithiobacillus Caldus (Aca) Short Prokaryotic Argonautes, Hnh-Associated (Sparha) With Grna And Tdna
Organism: Acidithiobacillus caldus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: IMMUNE SYSTEM/DNA/RNA Ligands: MN |
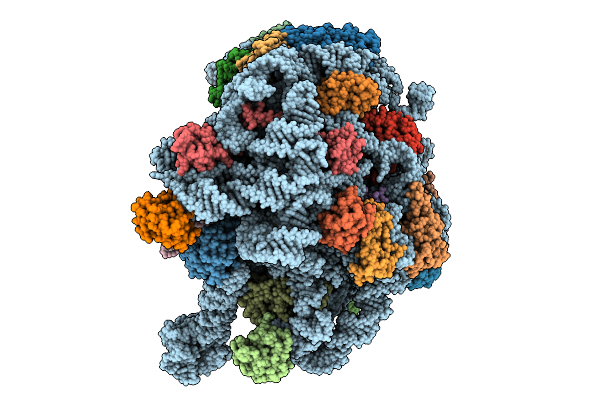 |
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME Ligands: MG, ZN |
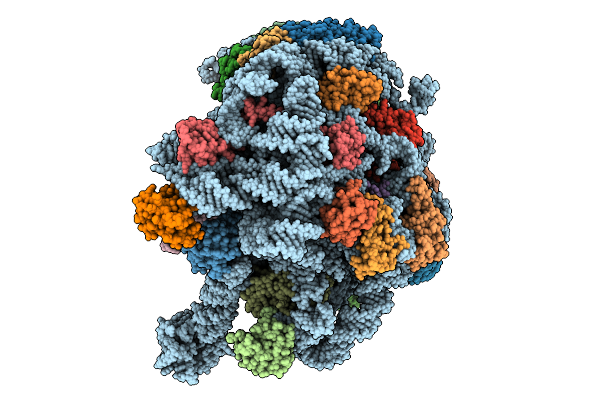 |
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME Ligands: MG, K, ZN |
 |
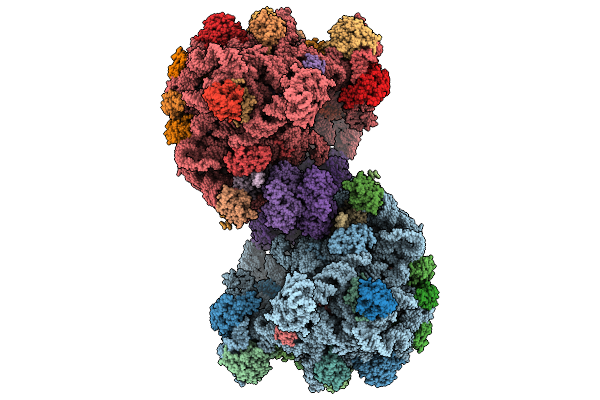
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME |
 |
Structure Of A Native Drosophila Melanogaster Pol Ii Elongation Complex Without Rpb4/Rpb7 Stalk
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Resolution:3.37 Å Release Date: 2025-05-14 Classification: TRANSCRIPTION Ligands: ZN |
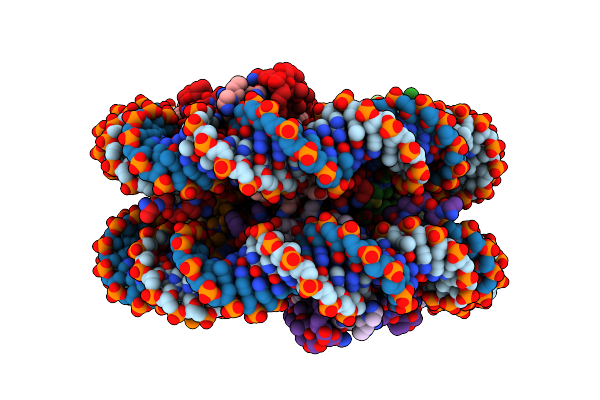 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: GENE REGULATION |
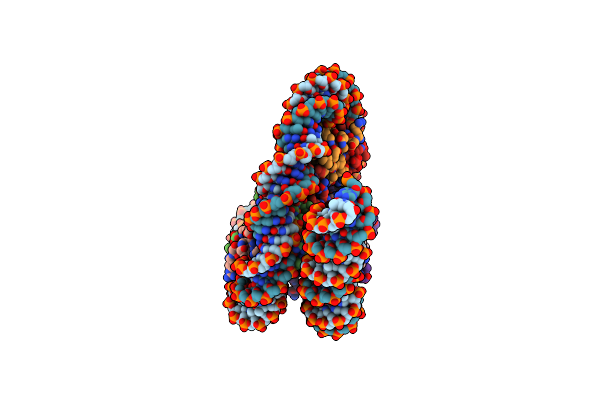 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: GENE REGULATION |
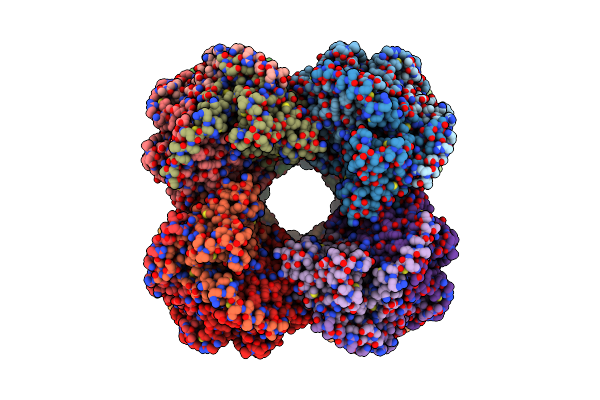 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: GENE REGULATION |
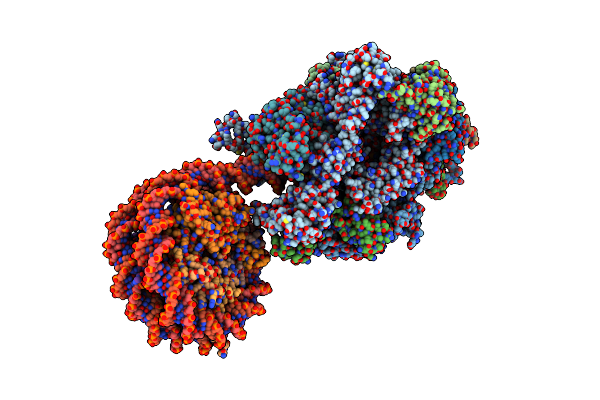 |
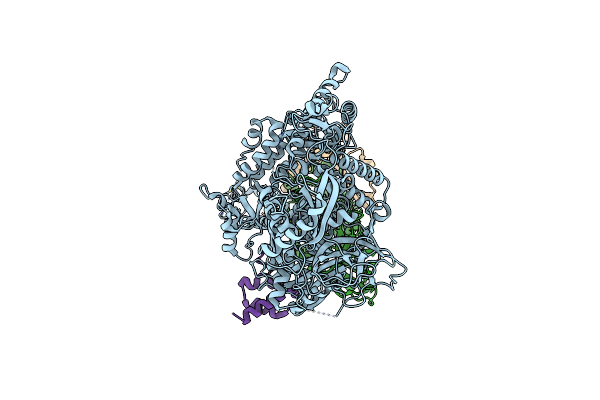
Structure Of A Native Drosophila Melanogaster Nucleosome Elongation Complex (Pol Ii Ec-Nucleosome). Composite Map
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: REPLICATION Ligands: ZN, SF4 |
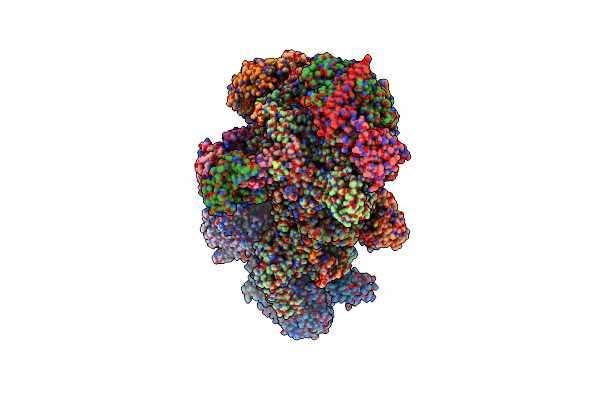 |
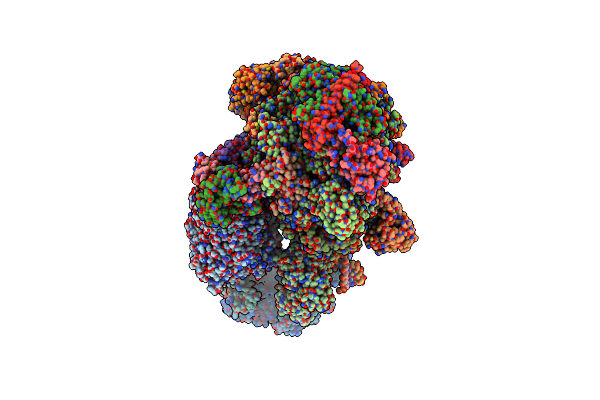
Method: ELECTRON MICROSCOPY
Release Date: 2024-10-30 Classification: TRANSCRIPTION Ligands: ZN, MG, SF4 |
 |
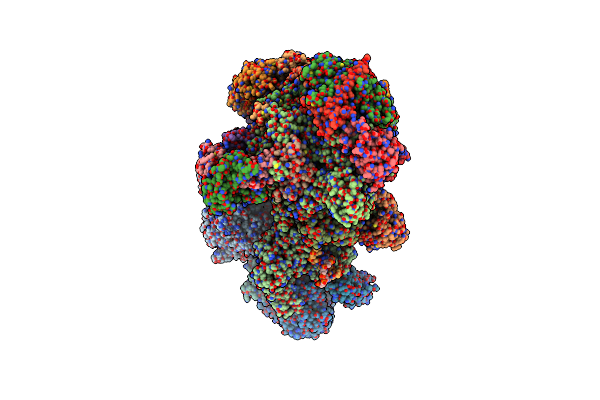
Method: ELECTRON MICROSCOPY
Release Date: 2024-10-30 Classification: TRANSCRIPTION Ligands: SF4, ZN, MG |
 |
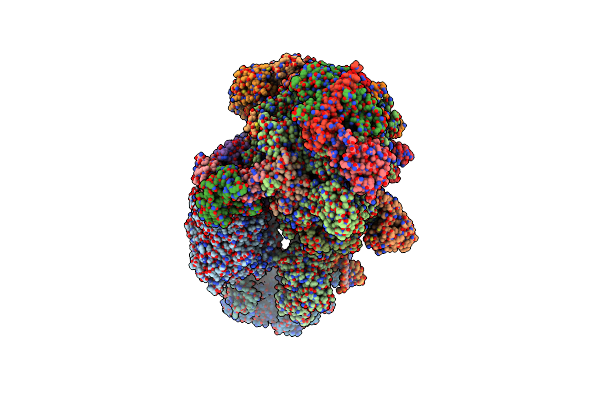
Method: ELECTRON MICROSCOPY
Release Date: 2024-10-23 Classification: TRANSCRIPTION Ligands: ZN, MG, SF4 |
 |
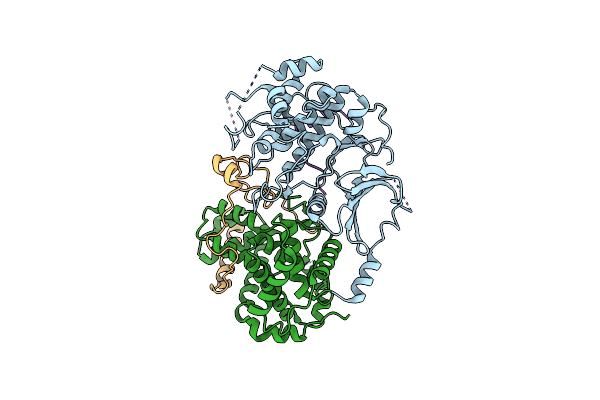
Method: ELECTRON MICROSCOPY
Release Date: 2024-10-23 Classification: TRANSCRIPTION Ligands: SF4, ZN, MG |
 |
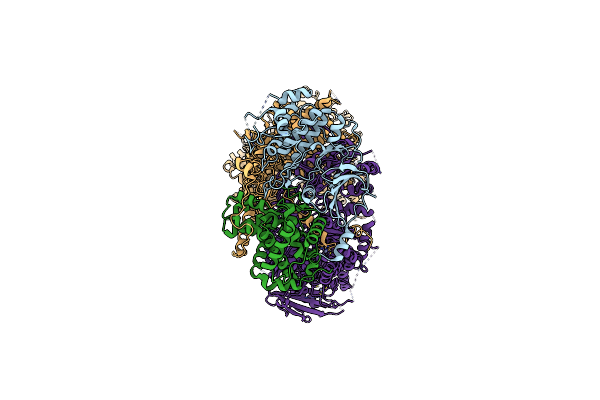
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: TRANSCRIPTION Ligands: ZN |
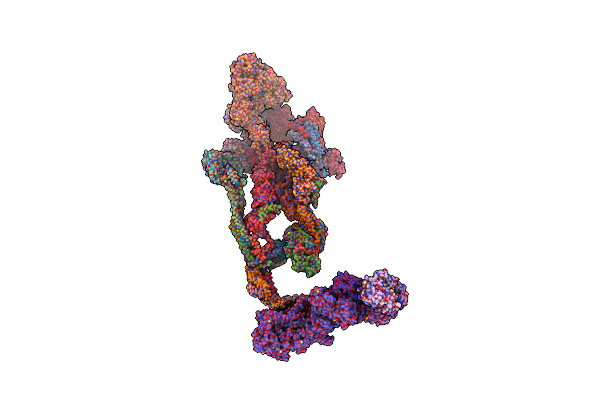 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: CHAPERONE |

