Search Count: 26
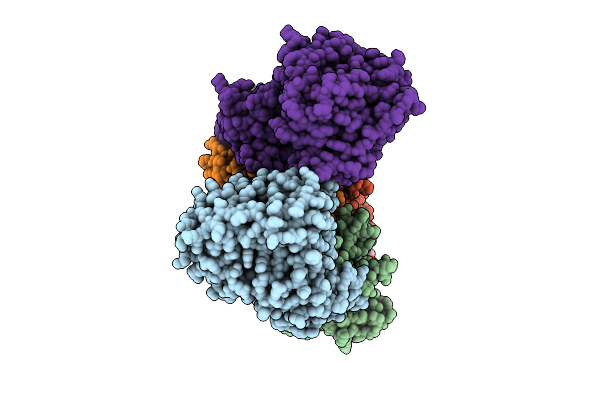 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, In The Absence Of Na+, Middle State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, CA, LMT, FES, FAD |
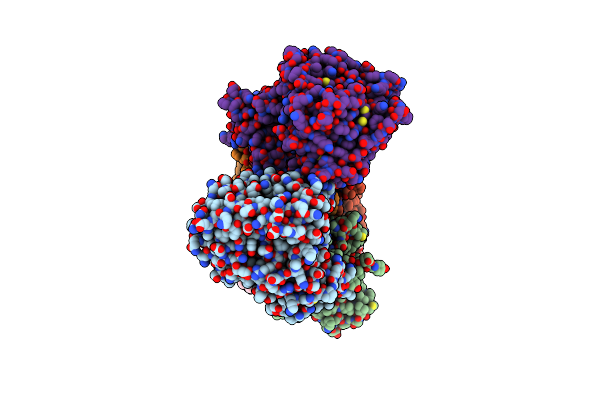 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrc-T225Y Mutant From Vibrio Cholerae
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, CA, FES, FAD |
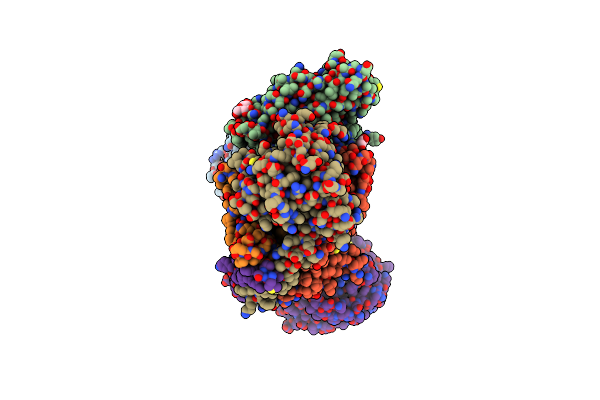 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrc-T225Y Mutant From Vibrio Cholerae Reduced By Nadh
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, CA, FES, FAD |
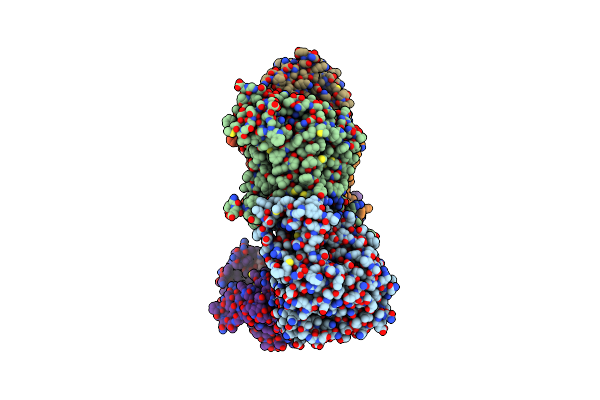 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-T236Y Mutant From Vibrio Cholerae
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: RBF, LMT, FMN, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-T236Y Mutant From Vibrio Cholerae Reduced By Nadh
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: RBF, LMT, CA, FMN, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, With Bound Korormicin A
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, IQT, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, In The Absence Of Na+, Upper State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, In The Absence Of Na+, Down State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-G141A Mutant From Vibrio Cholerae Reduced By Nadh, With Bound Korormicin A, Stable State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, IQT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-G141A Mutant From Vibrio Cholerae Reduced By Nadh, With Bound Korormicin A, Shifted State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, PEE, LMT, IQT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, With Bound Aurachin D-42
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, 0NI, PEE, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, CA, FES, FAD, NAI |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-G141A Mutant From Vibrio Cholerae With Bound Korormicin A
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:2.68 Å Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, PEE, IQT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Pumping Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae, State 1
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2022-07-20 Classification: TRANSLOCASE Ligands: FMN, RBF, PEE, LMT, FES, CA, FAD |
 |
Cryo-Em Structure Of Na+-Pumping Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae, State 2
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2022-07-20 Classification: TRANSLOCASE Ligands: FMN, RBF, PEE, LMT, FES, CA, FAD |
 |
Cryo-Em Structure Of Na+-Pumping Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae, State 3
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2022-07-20 Classification: TRANSLOCASE Ligands: FMN, RBF, PEE, LMT, FES, CA, FAD |
 |
Cryo-Em Structure Of Na+-Pumping Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae, With Aurachin D-42
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2022-07-20 Classification: TRANSLOCASE Ligands: FMN, RBF, 0NI, PEE, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Pumping Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae, With Korormicin
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2022-07-20 Classification: OXIDOREDUCTASE Ligands: FMN, RBF, IQT, PEE, LMT, CA, FES, FAD |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN Ligands: SF4, PC1, FES, FMN, 3PE, 88I, CDL, ATP, NDP, ZN, EHZ |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2014-12-03 Classification: TRANSFERASE/PROTEIN BINDING |

