Search Count: 9
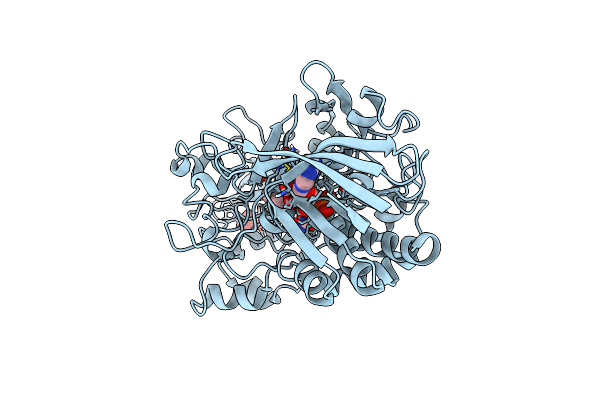 |
Crystal Structure And Charge Density Studies Of Cholesterol Oxidase From Brevibacterium Sterolicum At 0.74 Ultra-High Resolution
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Resolution:0.74 Å Release Date: 2015-04-15 Classification: OXIDOREDUCTASE Ligands: FAD, PO4 |
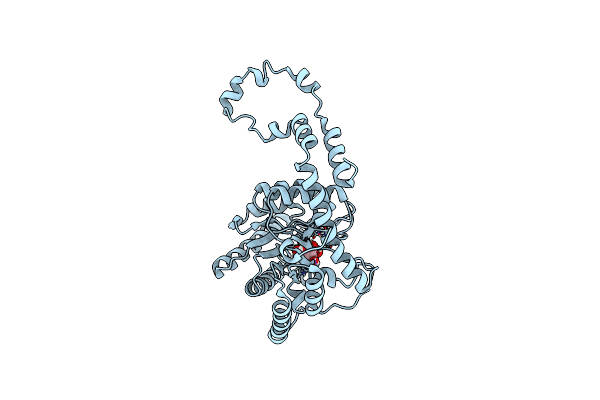 |
Neutron Structure Of The Cyclic Glucose Bound Xylose Isomerase E186Q Mutant
Organism: Streptomyces rubiginosus
Method: NEUTRON DIFFRACTION Resolution:2.1900 Å, 1.8400 Å Release Date: 2014-02-12 Classification: ISOMERASE Ligands: GLC, MN, MG, DOD |
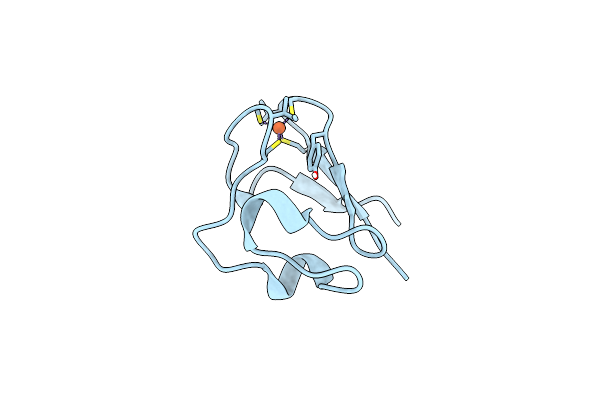 |
Neutron Structure Of Perdeuterated Rubredoxin Refined Against 1.75 Resolution Data Collected On The New Imagine Instrument At Hfir, Ornl
Organism: Pyrococcus furiosus
Method: NEUTRON DIFFRACTION Resolution:1.75 Å Release Date: 2013-12-04 Classification: ELECTRON TRANSPORT Ligands: FE, DOD |
 |
The X-Ray Crystal Structure Of A Thermophilic Cellobiose Binding Protein Bound With Laminaribiose
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2013-10-09 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
The X-Ray Crystal Structure Of A Thermophilic Cellobiose Binding Protein Bound With Laminaripentaose
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2013-10-09 Classification: SUGAR BINDING PROTEIN Ligands: CA |
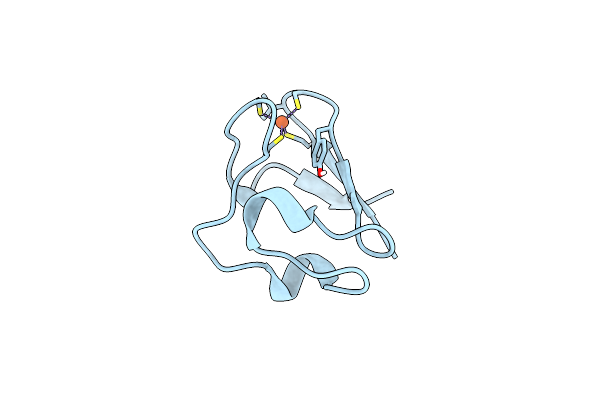 |
Organism: Pyrococcus furiosus
Method: NEUTRON DIFFRACTION Resolution:1.75 Å Release Date: 2011-12-28 Classification: ELECTRON TRANSPORT Ligands: FE, DOD |
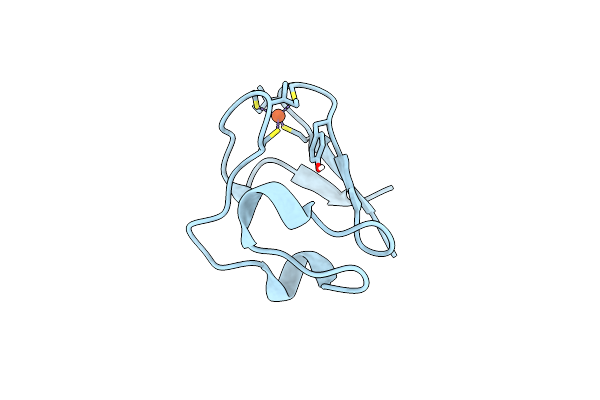 |
Organism: Pyrococcus furiosus
Method: NEUTRON DIFFRACTION Resolution:1.75 Å Release Date: 2011-12-28 Classification: ELECTRON TRANSPORT Ligands: FE, DOD |
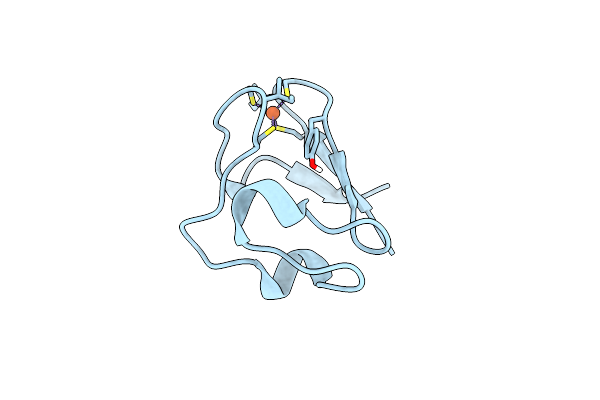 |
Organism: Pyrococcus furiosus
Method: NEUTRON DIFFRACTION Resolution:1.75 Å Release Date: 2011-12-28 Classification: ELECTRON TRANSPORT Ligands: FE, DOD |
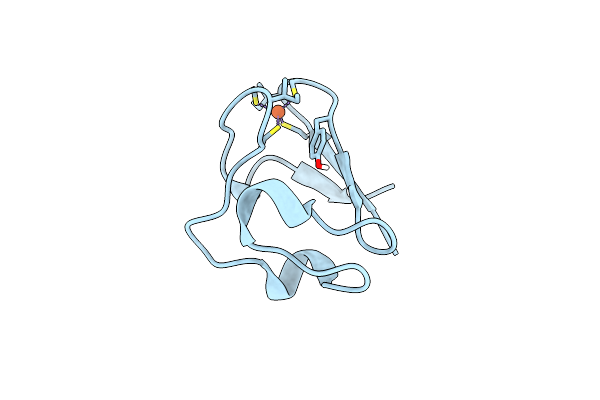 |
Organism: Pyrococcus furiosus dsm 3638
Method: NEUTRON DIFFRACTION Resolution:1.75 Å Release Date: 2011-12-28 Classification: ELECTRON TRANSPORT Ligands: FE, DOD |

