Search Count: 20
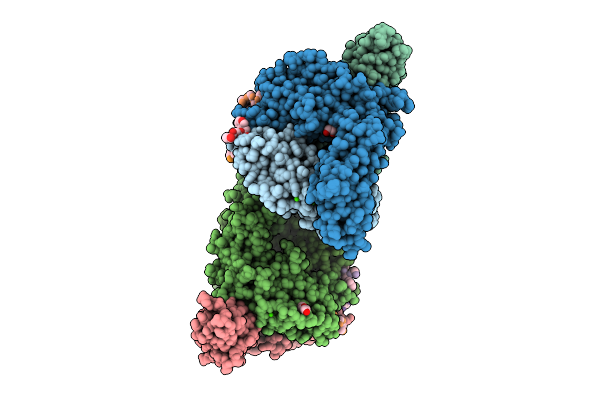 |
Vibrio Cholerae Protein Frha Peptid-Binding Domain And Adjacent Split Domain (S1127-F1439) In Complex With Peptide Agwtd X-Ray Crystallography Structure
Organism: Vibrio cholerae o395, Vibrio cholerae
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: METAL BINDING PROTEIN Ligands: CA, PEG, EDO |
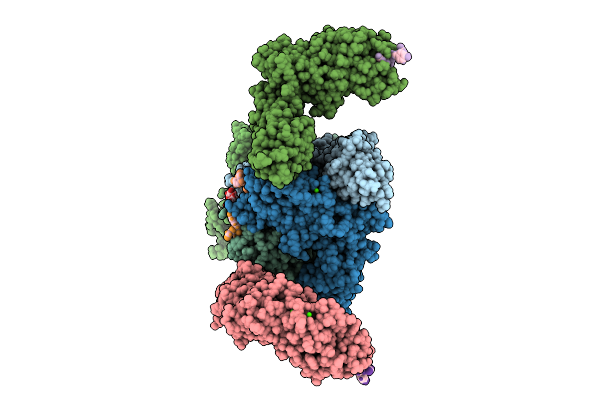 |
Vibrio Cholerae Protein Frha Peptid-Binding Domain And Adjacent Split Domain (S1127-F1439) In Complex With Peptide Agytd X-Ray Crystallography Structure
Organism: Vibrio cholerae o395, Vibrio cholerae
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: METAL BINDING PROTEIN Ligands: CA |
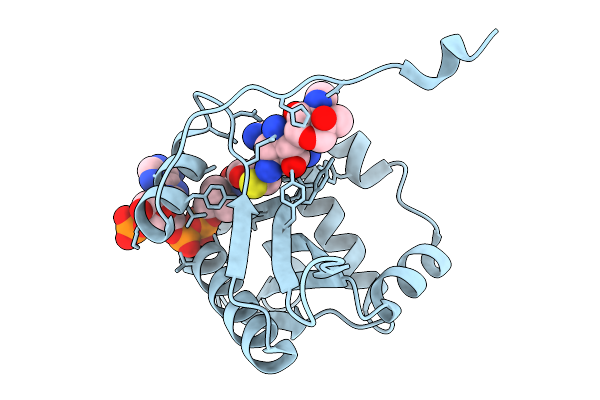 |
Structure Of Aminoglycoside Acetyltransferase Aac(3)-Ia In Complex With Sisomicin And Coa
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: COA, SIS |
 |
Structure Of N-3 Aminoglycoside Acetyltransferase Xia (Aac(3)-Xia) In Complex With Coenzyme A And Tobramycin
Organism: Corynebacterium striatum
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: COA, TOY |
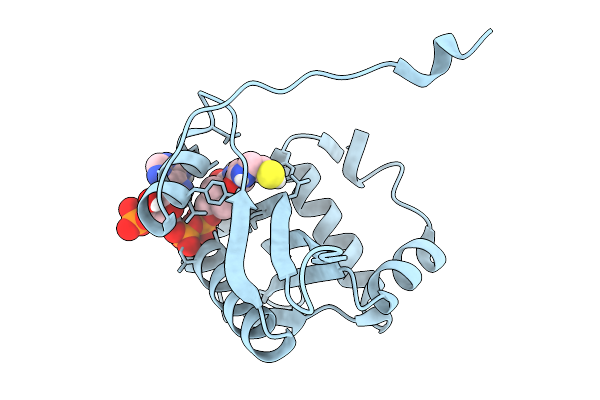 |
Structure Of Aminoglycoside Acetyltransferase Aac(3)-Ia In Complex With Coa
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: COA |
 |
Structure Of N-3 Aminoglycoside Acetyltransferase Xi (Aac(3)-Xi) In Complex With Acetyl Coenzyme A
Organism: Corynebacterium striatum
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: ACO, PEG, GOL |
 |
Structure Of N-3 Aminoglycoside Acetyltransferase Xi (Aac(3)-Xi) In Complex With Coenzyme A And Acetylated Sisomicin
Organism: Corynebacterium striatum
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: COA, DVM, ACO |
 |
Structure Of N-3 Aminoglycoside Acetyltransferase Xia (Aac(3)-Xia) In Complex With Acetylamino Coenzyme A And Gentamicin
Organism: Corynebacterium striatum
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: 51G, 6IM, ACT |
 |
Structure Of N-3 Aminoglycoside Acetyltransferase Xi (Aac(3)-Xi) In Complex With Coenzyme A, And One Apo Monomer
Organism: Corynebacterium striatum
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: ACO, GOL, PO4 |
 |
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-11-20 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, JQG, A1AMO, MN |
 |
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-11-20 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, A1AMN, MN |
 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2024-09-04 Classification: MOTOR PROTEIN |
 |
Organism: Stanieria sp. nies-3757
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: LIGASE |
 |
Cryo-Em Structure Of Stanieria Sp. Cpha2 In Complex With Adpcp And 4X(Beta-Asp-Arg)
Organism: stanieria sp. nies-3757, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: LIGASE Ligands: YHZ, MG, ACP |
 |
Crystal Structure Of The M. Abscessus Leurs Editing Domain In Complex With Epetraborole-Amp Adduct
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-10-06 Classification: LIGASE/LIGASE INHIBITOR Ligands: SO4 |
 |
Crystal Structure Of The M. Abscessus Leurs Editing Domain In Complex With Epetraborole-Amp Adduct
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-10-06 Classification: LIGASE/LIGASE INHIBITOR Ligands: 365, SO4 |
 |
Organism: Arabidopsis thaliana, Glycine max
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2019-03-27 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2014-11-12 Classification: TRANSCRIPTION/TRANSFERASE |
 |
Solution Structure Of The Small Dictyostelium Discoideium Myosin Light Chain Mlcb Provides Insights Into Iq-Motif Recognition Of Class I Myosin Myo1B
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2013-12-11 Classification: CONTRACTILE PROTEIN |
 |

