Search Count: 20
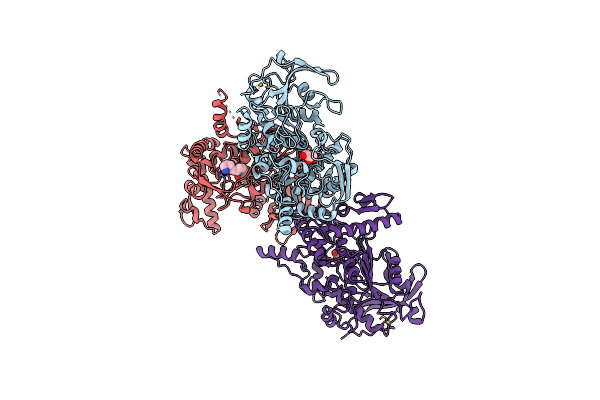 |
Organism: Fusobacterium nucleatum subsp. nucleatum atcc 25586
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-03-24 Classification: HYDROLASE Ligands: GOL, MG, SPD |
 |
Crystal Structure Of Native 6-Phospho-Glucosidase Lpbgl From Lactobacillus Plantarum
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2017-08-02 Classification: HYDROLASE Ligands: PO4 |
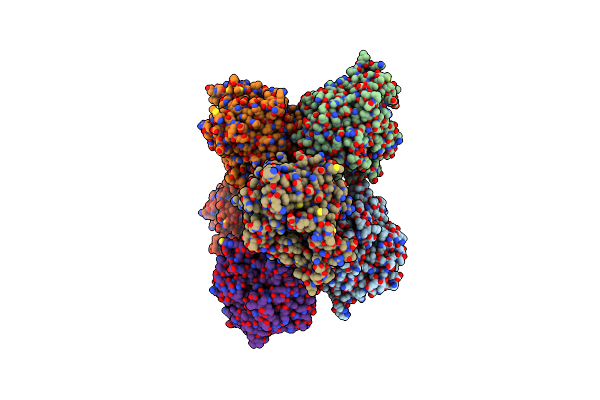 |
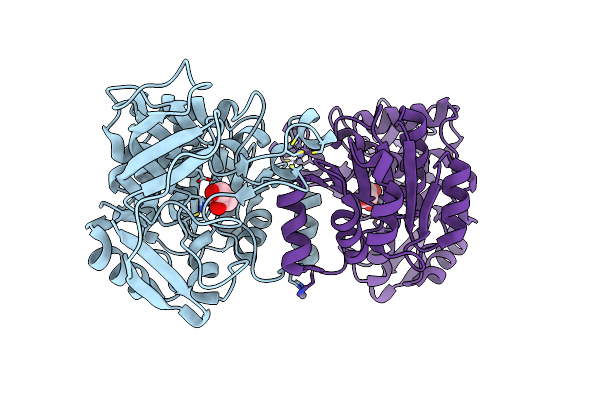
Crystal Structure Of The Double Mutant (Cys211Ser/Cys292Ser) 6-Phospho-B-D-Glucosidase From Lactobacillus Plantarum
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-08-02 Classification: HYDROLASE |
 |
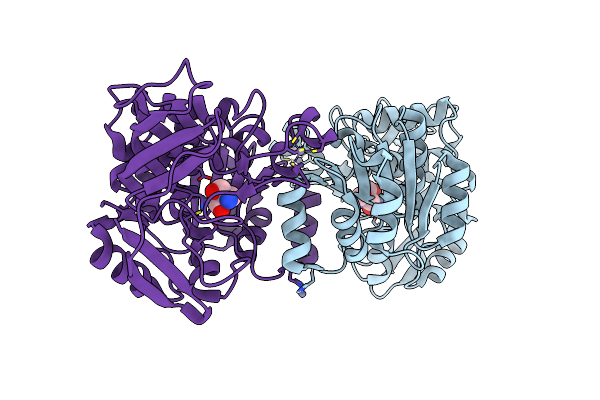
Closed State Of Galactitol-1-Phosphate 5-Dehydrogenase From E. Coli In Complex With Glycerol.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2015-07-15 Classification: OXIDOREDUCTASE Ligands: ZN, GOL |
 |
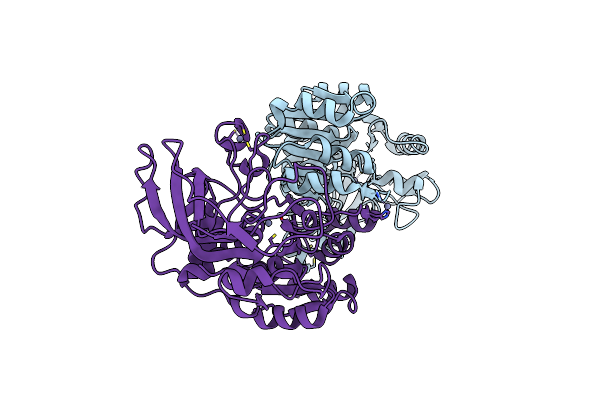
Galactitol-1-Phosphate 5-Dehydrogenase From E. Coli With Tris Within The Active Site.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-07-15 Classification: OXIDOREDUCTASE Ligands: ZN, TRS |
 |
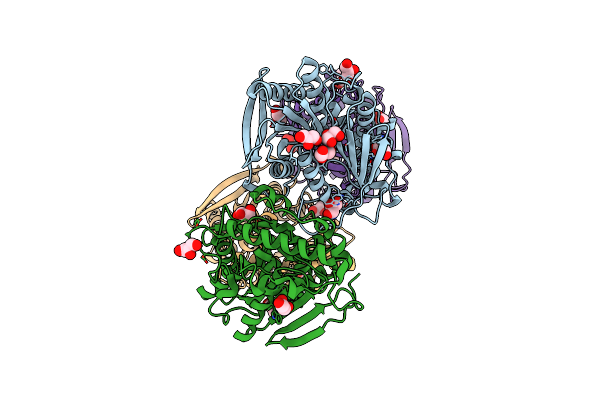
Open State Of Galactitol-1-Phosphate 5-Dehydrogenase From E. Coli, With Zinc In The Catalytic Site.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-07-15 Classification: OXIDOREDUCTASE Ligands: ZN |
 |
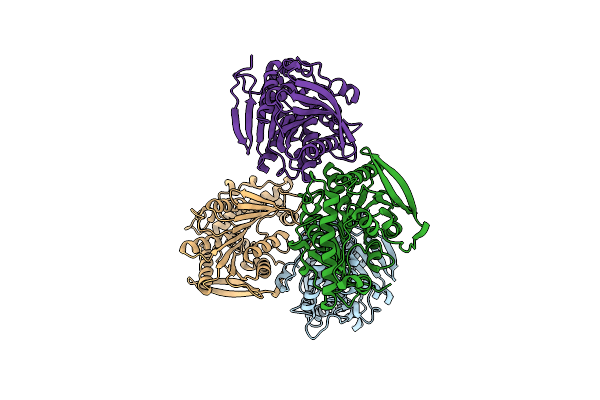
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2014-04-02 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2014-04-02 Classification: HYDROLASE |
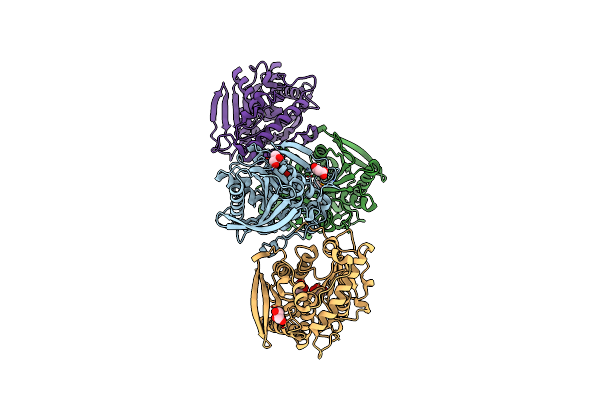 |
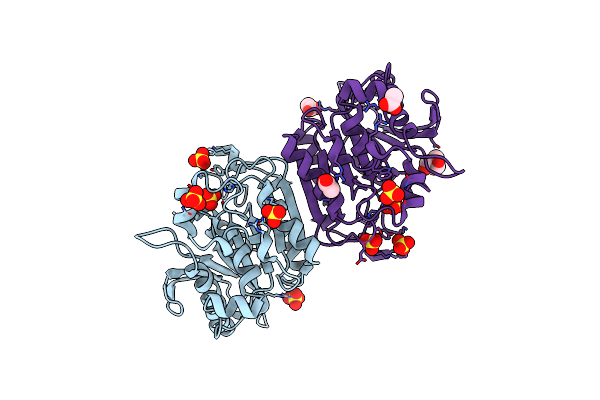
Crystal Structure Of Carboxylesterase Lpest1 From Lactobacillus Plantarum: High Resolution Model
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2014-04-02 Classification: HYDROLASE Ligands: GOL, MLI |
 |
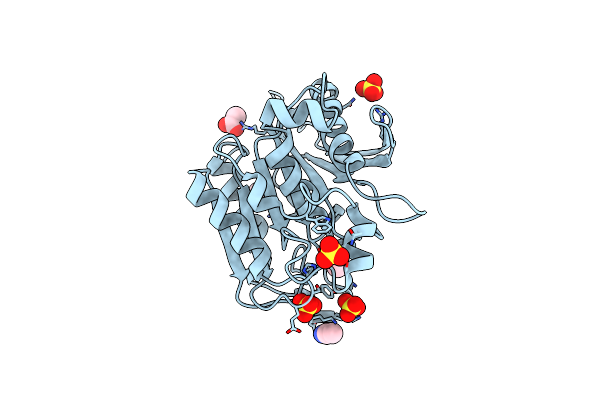
Complete Crystal Structure Of The Carboxylesterase Cest-2923 (Lp_2923) From Lactobacillus Plantarum Wcfs1
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2013-10-30 Classification: HYDROLASE Ligands: SO4, ACT, 144 |
 |
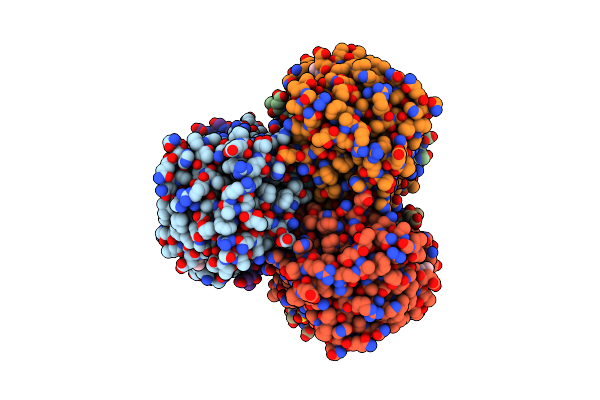
Complete Crystal Structure Of Carboxylesterase Cest-2923 From Lactobacillus Plantarum Wcfs1
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2013-10-30 Classification: HYDROLASE Ligands: ACT, SO4, CCN |
 |
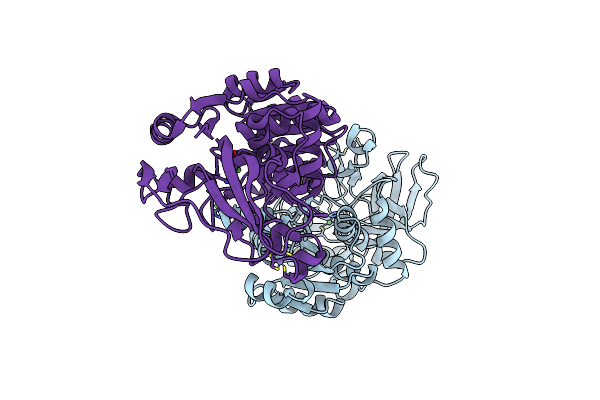
Complete Crystal Structure Of Carboxylesterase Cest-2923 (Lp_2923) From Lactobacillus Plantarum Wcfs1
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-10-30 Classification: HYDROLASE Ligands: ACT, SO4, QY9, CCN |
 |
Crystal Structure Of Galactitol-1-Phosphate Dehydrogenase From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2012-08-08 Classification: OXIDOREDUCTASE Ligands: NI, ZN |
 |
High-Resolution Structural Insights On The Sugar-Recognition And Fusion Tag Properties Of A Versatile B-Trefoil Lectin Domain
Organism: Laetiporus sulphureus
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2011-10-12 Classification: SUGAR BINDING PROTEIN |
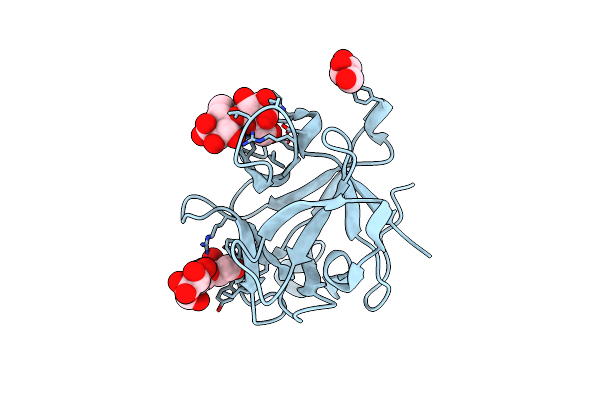 |
High-Resolution Structural Insights On The Sugar-Recognition And Fusion Tag Properties Of A Versatile B-Trefoil Lectin Domain
Organism: Laetiporus sulphureus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2011-10-12 Classification: SUGAR BINDING PROTEIN Ligands: GOL |
 |
Crystal Structure Of Single Point Mutant Arg48Gln Of P-Coumaric Acid Decarboxylase From Lactobacillus Plantarum Structural Insights Into The Active Site And Decarboxylation Catalytic Mechanism
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2010-02-09 Classification: LYASE Ligands: BA |
 |
Crystal Structure Of Single Point Mutant Glu71Ser P-Coumaric Acid Decarboxylase
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2010-02-09 Classification: LYASE Ligands: BA, IPA, NA, CL |
 |
Crystal Structure Of P-Coumaric Acid Decarboxylase From Lactobacillus Plantarum: Structural Insights Into The Active Site And Decarboxylation Catalytic Mechanism
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2009-11-17 Classification: LYASE |
 |
Crystal Structure Of Single Point Mutant Tyr20Phe P-Coumaric Acid Decarboxylase From Lactobacillus Plantarum: Structural Insights Into The Active Site And Decarboxylation Catalytic Mechanism
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2009-11-17 Classification: LYASE Ligands: ACT, IPA |
 |
Crystal Structure Of The Hexameric Catabolic Ornithine Transcarbamylase From Lactobacillus Hilgardii
Organism: Lactobacillus hilgardii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-11-17 Classification: TRANSFERASE Ligands: NI |

