Search Count: 37
 |
Crystal Structure Of Catalytic Domain Of Lytb (E585Q) From Streptococcus Pneumoniae In Complex With Nag-Nam-Nag-Nam Tetrasaccharide
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-09-21 Classification: HYDROLASE Ligands: 1PE, PEG, CA |
 |
Crystal Structure Of Catalytic Domain In Open Conformation Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PGE, PEG, CA |
 |
Crystal Structure Of Catalytic Domain In Closed Conformation Of Lytb (E585Q)From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PGE, PEG, ACT, CA |
 |
Crystal Structure Of Catalytic Domain Of Lytb From Streptococcus Pneumoniae In Complex With Nag-Nag-Nag-Nag Tetrasaccharide
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PEG, CA |
 |
Crystal Structure Of Catalytic Domain Of Lytb (E585Q) From Streptococcus Pneumoniae In Complex With Nag-Nam-Nag-Nam-Nag Peptidolycan Analogue
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PGE, PEG, CA |
 |
Crystal Structure Of Choline-Binding Module Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: CHT |
 |
Crystal Structure Of Catalytic Domain In Closed Conformation Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PEG, CA |
 |
Crystal Structure Of Choline-Binding Module (R1-R9) Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: CHT, ZN, PGE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2022-06-22 Classification: ONCOPROTEIN Ligands: 0LI, TRS, GOL |
 |
Crystal Structure Of Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: EPE, CD, CL |
 |
Crystal Structure Of Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus In Complex With Piperacillin
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: YPP |
 |
Crystal Structure Of Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus In Complex With Penicillin G
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: CIT, SO4, PNM |
 |
Crystal Structure Of Pasta Domains Of The Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus In Complex With Pentaglycine
Organism: Staphylococcus aureus (strain col), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.36 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: CD, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2018-05-30 Classification: TRANSFERASE Ligands: AGS |
 |
Crystal Structure Of The Essential Protein Pcsb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2014-05-21 Classification: CELL CYCLE Ligands: EDO, CL, MG, PG4 |
 |
The Crystal Structure Of The Ing4 Dimerization Domain Reveals The Functional Organization Of The Ing Family Of Chromatin Binding Proteins.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2012-02-22 Classification: CELL CYCLE |
 |
Crystal Structure Of The Mammalian Cytosolic Chaperonin Cct In Complex With Tubulin
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:5.50 Å Release Date: 2010-12-15 Classification: CHAPERONE |
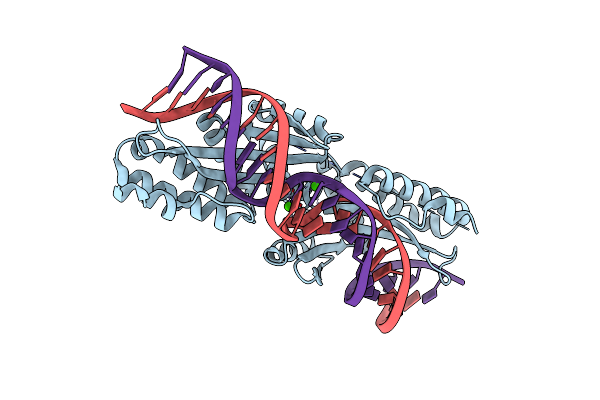 |
Molecular Basis Of Engineered Meganuclease Targeting Of The Endogenous Human Rag1 Locus
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-10-06 Classification: HYDROLASE/DNA Ligands: CA |
 |
Molecular Basis Of Engineered Meganuclease Targeting Of The Endogenous Human Rag1 Locus
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-10-06 Classification: HYDROLASE/DNA Ligands: MN, EOH, 1PE |

