Search Count: 55
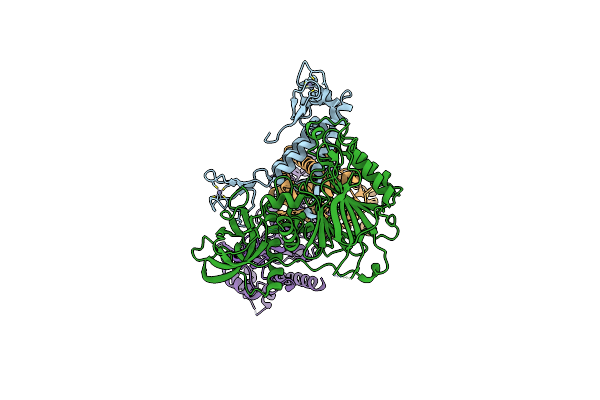 |
Tetramer Core Subcomplex (Conformation 1) Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: REPLICATION, TRANSFERASE Ligands: ZN |
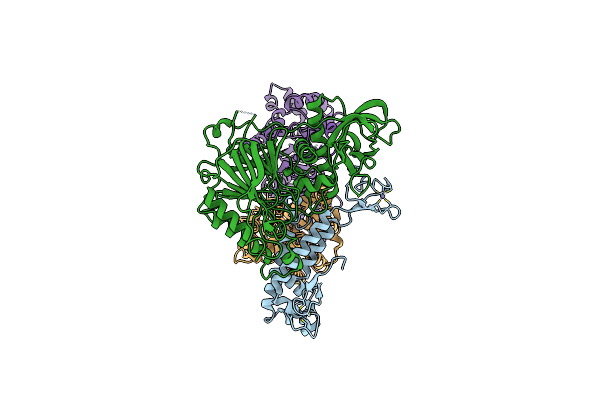 |
Tetramer Core Subcomplex (Conformation 2) Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: REPLICATION, TRANSFERASE Ligands: ZN |
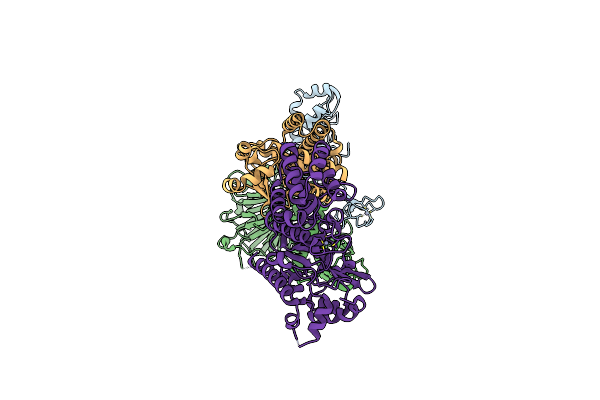 |
Tetramer Core Subcomplex (Conformation 3) Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: REPLICATION, TRANSFERASE Ligands: ZN |
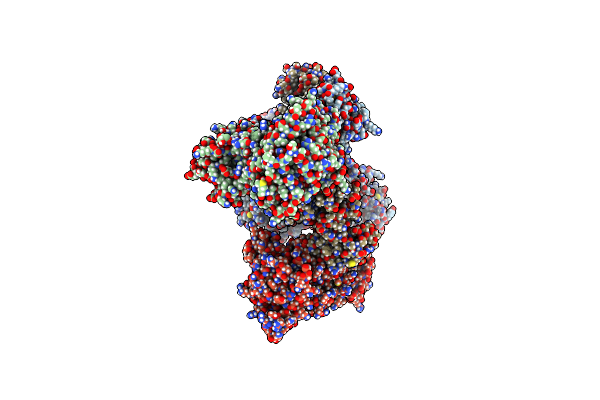 |
Dna Initiation Complex (Configuration 1) Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: REPLICATION, TRANSFERASE Ligands: ZN, DGT, MG, SF4 |
 |
Dna Initiation Complex (Configuration 2) Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: ZN, DGT, MG, SF4 |
 |
Dna Elongation Complex (Configuration 1) Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: ZN, DGT, MG, SF4 |
 |
Dna Elongation Complex (Configuration 2) Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: ZN, DGT, MG, SF4 |
 |
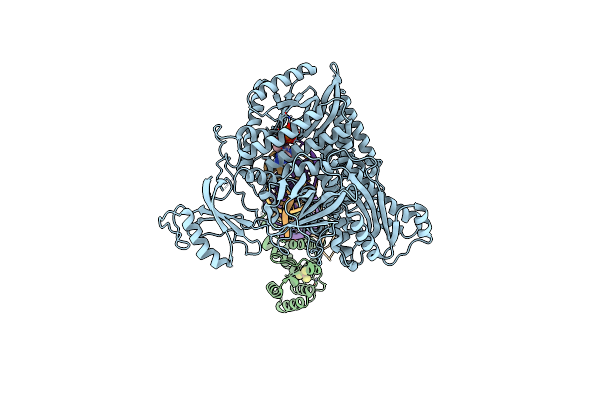
Partial Dna Termination Subcomplex Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: DGT, MG |
 |
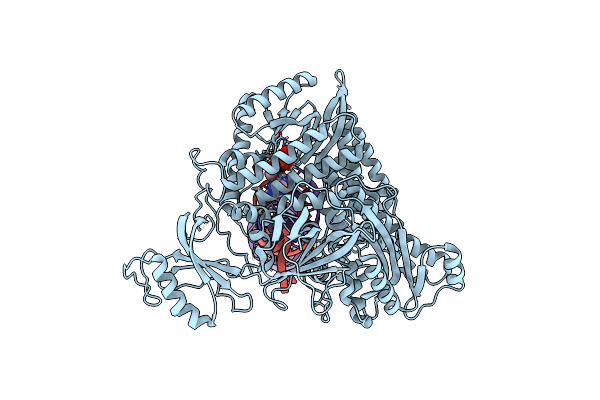
Complete Dna Termination Subcomplex 1 Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: DGT, MG, SF4 |
 |
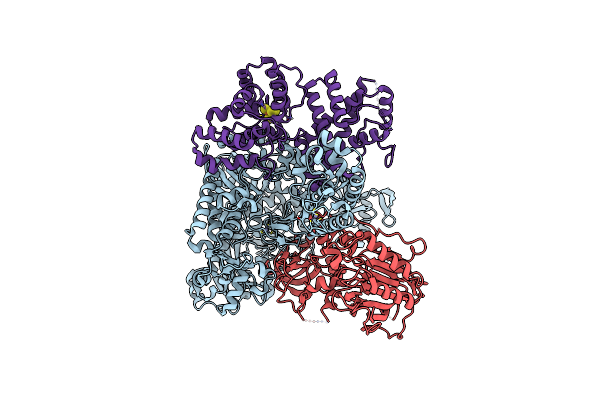
Complete Dna Termination Subcomplex 2 Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: DGT, MG |
 |
Partial Auto-Inhibitory Complex Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: REPLICATION, TRANSFERASE Ligands: ZN, SF4 |
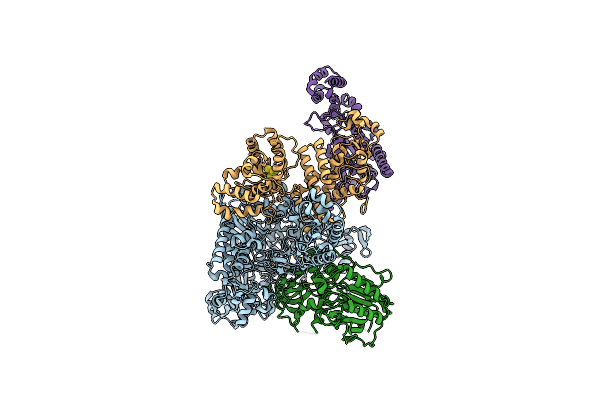 |
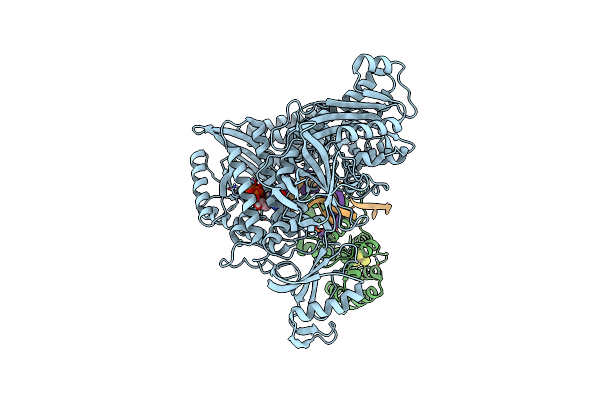
Complete Auto-Inhibitory Complex Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: REPLICATION, TRANSFERASE Ligands: ZN, SF4 |
 |
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: DGT, MG, SF4 |
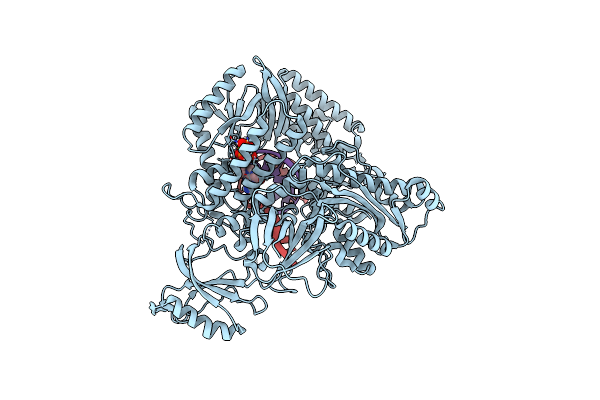 |
Partial Dna Elongation Subcomplex Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: DGT, MG |
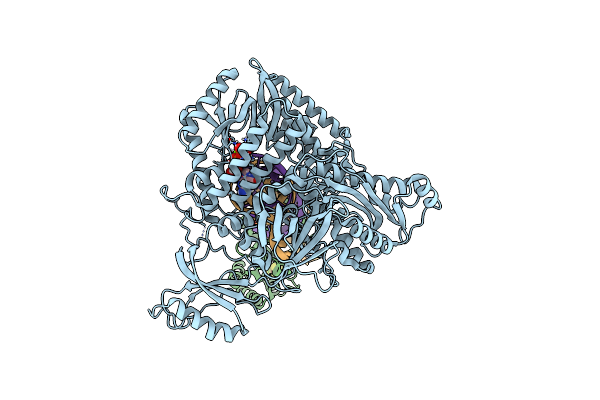 |
Complete Dna Elongation Subcomplex Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: DGT, MG, SF4 |
 |
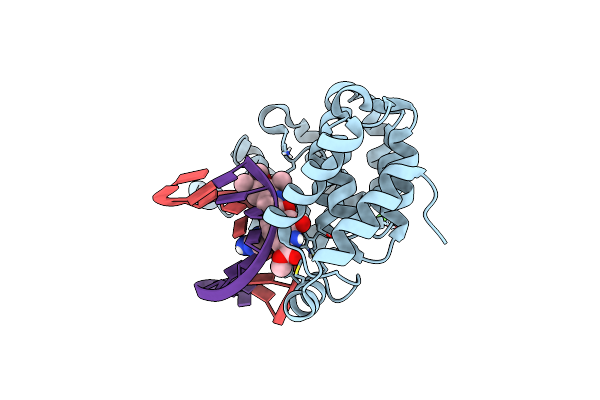
Bacillus Cereus Dna Glycosylase Alkd Bound To A Cc1065-Adenine Nucleobase Adduct And Dna Containing An Abasic Site
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2021-11-17 Classification: HYDROLASE/DNA/ANTIBIOTIC Ligands: CA, YNG |
 |
Bacillus Cereus Dna Glycosylase Alkd Bound To A Duocarmycin Sa-Adenine Nucleobase Adduct And Dna Containing An Abasic Site
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2021-11-17 Classification: HYDROLASE/DNA/ANTIBIOTIC Ligands: CA, YNY |
 |
Pseudomonas Fluorescens Alkylpurine Dna Glycosylase Alkc Bound To Dna Containing An Oxocarbenium-Intermediate Analog
Organism: Pseudomonas fluorescens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-10-25 Classification: HYDROLASE/DNA Ligands: GOL, MES, NA, 9B4 |
 |
Pseudomonas Fluorescens Alkylpurine Dna Glycosylase Alkc Bound To Dna Containing An Abasic Site Analog
Organism: Pseudomonas fluorescens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-10-25 Classification: HYDROLASE/DNA Ligands: P4K, NA |
 |
Bacillus Cereus Dna Glycosylase Alkd Bound To A Yatakemycin-Adenine Nucleobase Adduct And Dna Containing An Abasic Site (12-Mer Product Complex)
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2017-07-19 Classification: HYDROLASE/DNA/ANTIBIOTIC Ligands: YTA |

