Search Count: 100
 |
Structure Of Native Leukocyte Myeloperoxidase In Complex With A Truncated Version Of The Staphylococcal Peroxidase Inhibitor Spin And Selenocyanate At Ph 5.5
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: SEK, HEM, NAG, CA |
 |
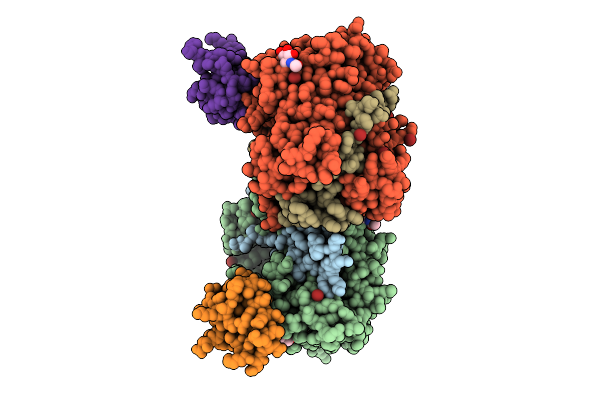
Structure Of Native Leukocyte Myeloperoxidase In Complex With A Truncated Version Of The Staphylococcal Peroxidase Inhibitor Spin And Thiocyanate At Ph 7.5
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: NAG, CA, SCN, HEM |
 |
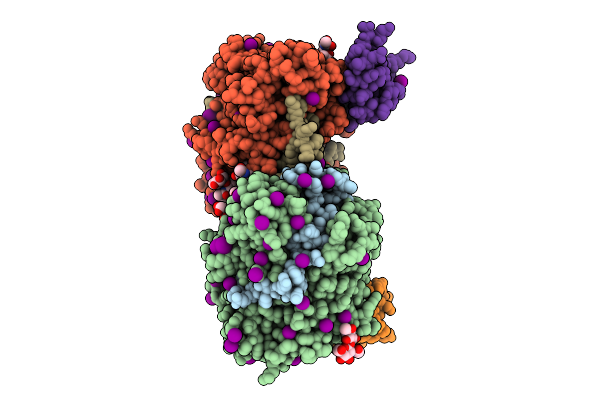
Structure Of Native Leukocyte Myeloperoxidase In Complex With A Truncated Version Of The Staphylococcal Peroxidase Inhibitor Spin And Bromide At Ph 7.5
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: HEM, BR, NAG, CA |
 |
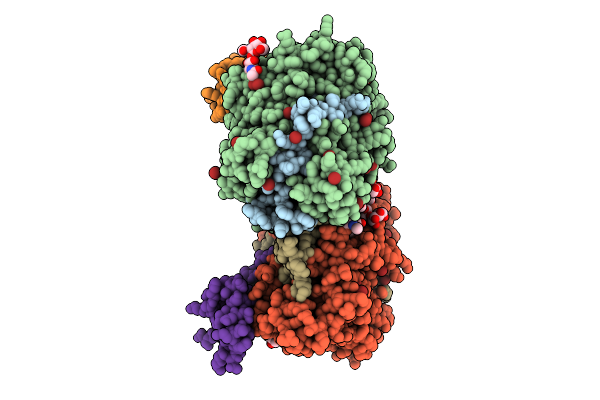
Structure Of Native Leukocyte Myeloperoxidase In Complex With A Truncated Version Of The Staphylococcal Peroxidase Inhibitor Spin And Iodide At Ph 5.5
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: IOD, CA, HEM |
 |
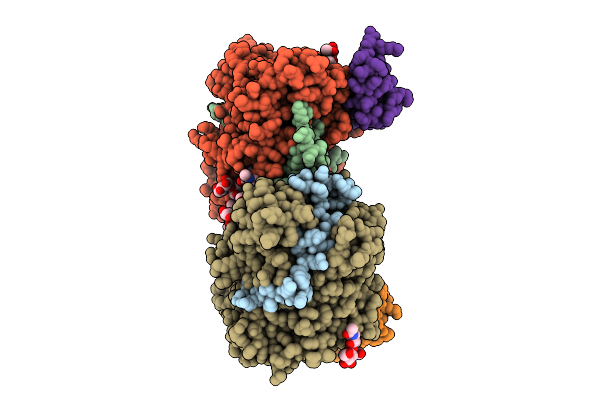
Structure Of Native Leukocyte Myeloperoxidase In Complex With The Staphylococcal Peroxidase Inhibitor Spin And Bromide At Ph 5.5
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: NAG, BR, CA, HEM |
 |
Structure Of Native Leukocyte Myeloperoxidase In Complex With A Truncated Version Of The Staphylococcal Peroxidase Inhibitor Spin And Chloride At Ph 5.5
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: HEM, IOD, CA, CL, NAG |
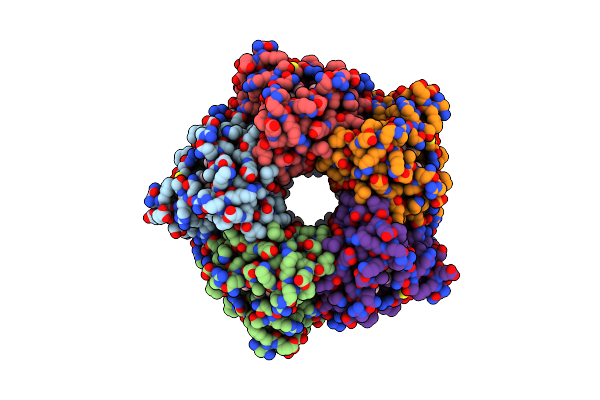 |
Cryo-Em Structure Of Coproheme Decarboxylase From Corynebacterium Diphtheriae In Complex With Heme B
Organism: Corynebacterium diphtheriae
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: HEM |
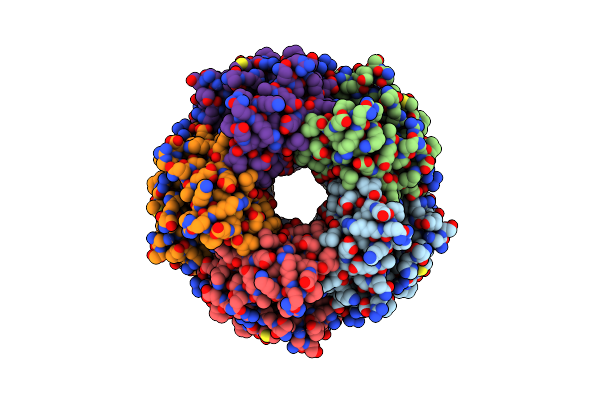 |
Cryo-Em Structure Of Apo Coproheme Decarboxylase From Corynebacterium Diphtheria.
Organism: Corynebacterium diphtheriae
Method: ELECTRON MICROSCOPY Resolution:2.27 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE |
 |
Coproporphyrin Iii - Lmcpfc Wt Complex Soaked With Fe2+ And Anomalous Densities
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-04-24 Classification: METAL BINDING PROTEIN Ligands: HT9, EDO, FE, CL |
 |
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-04-24 Classification: METAL BINDING PROTEIN Ligands: FEC, EDO, CA |
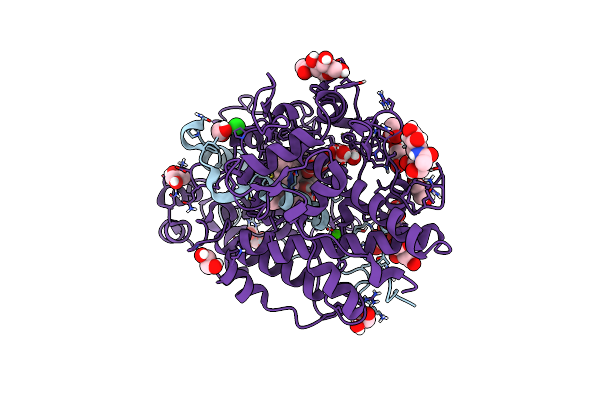 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-01-24 Classification: OXIDOREDUCTASE Ligands: HEB, GOL, EDO, FLC, CL, NAG, ACT, CA |
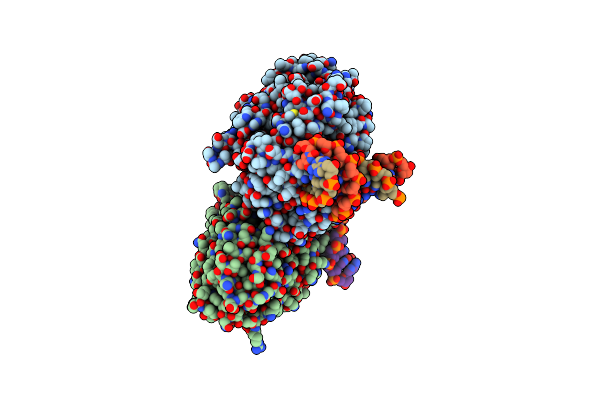 |
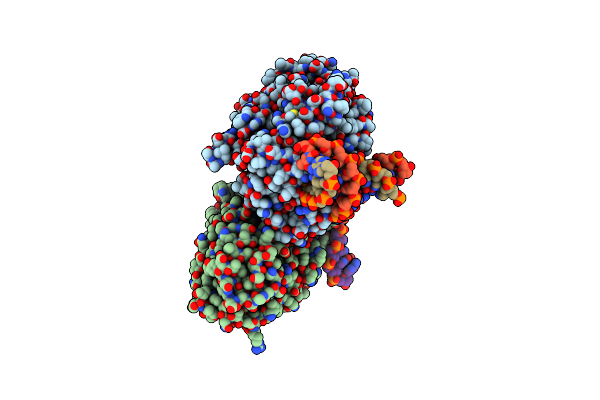
Dark, Fully Reduced Structure Of The Mmcpdii-Dna Complex As Produced At Swissfel
Organism: Methanosarcina mazei, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-11-22 Classification: DNA BINDING PROTEIN Ligands: FAD, SO4 |
 |
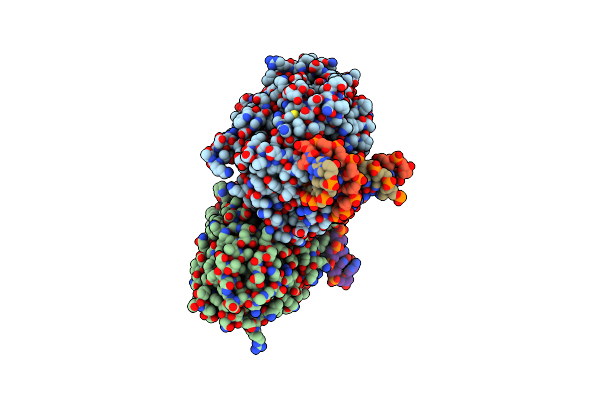
Tr-Sfx Mmcpdii-Dna Complex: 100 Ps Snapshot. Includes 100Ps, Dark, And Extrapolated Structure Factors
Organism: Methanosarcina mazei, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-22 Classification: DNA BINDING PROTEIN Ligands: SO4, FAD |
 |
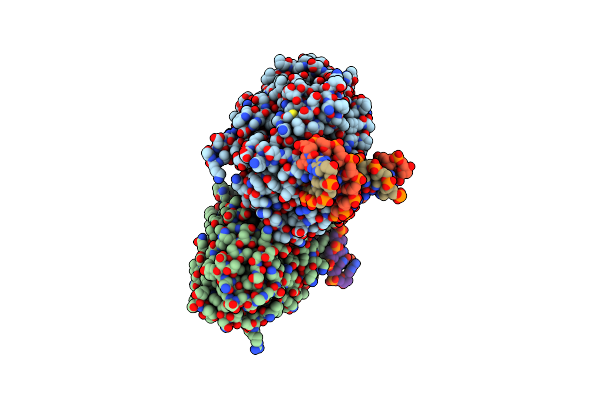
Tr-Sfx Mmcpdii-Dna Complex: 250 Ps Snapshot. Includes 250 Ps, Dark, And Extrapolated Structure Factors
Organism: Methanosarcina mazei, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2023-11-22 Classification: DNA BINDING PROTEIN Ligands: FAD, SO4 |
 |
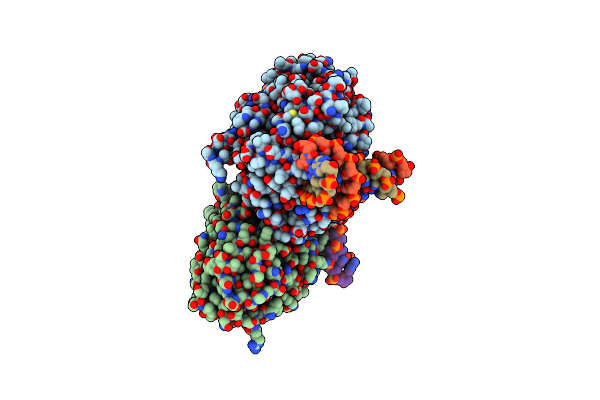
Tr-Sfx Mmcpdii-Dna Complex: 450 Ps Snapshot. Includes 450Ps, Dark, And Extrapolated Structure Factors
Organism: Methanosarcina mazei, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-11-22 Classification: DNA BINDING PROTEIN Ligands: FAD, SO4 |
 |
Tr-Sfx Mmcpdii-Dna Complex: 650 Ps Snapshot. Includes 650Ps, Dark, And Extrapolated Structure Factors
Organism: Methanosarcina mazei, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-11-22 Classification: DNA BINDING PROTEIN Ligands: FAD, SO4 |
 |
Tr-Sfx Mmcpdii-Dna Complex: 1 Ns Snapshot. Includes 1 Ns, Dark, And Extrapolated Structure Factors
Organism: Methanosarcina mazei, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-11-22 Classification: DNA BINDING PROTEIN Ligands: SO4, FAD |
 |
Tr-Sfx Mmcpdii-Dna Complex: 2 Ns Snapshot. Includes 2 Ns, Dark, And Extrapolated Structure Factors
Organism: Methanosarcina mazei, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-11-22 Classification: DNA BINDING PROTEIN Ligands: FAD, SO4 |
 |
Organism: Methanosarcina mazei, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2023-11-22 Classification: DNA BINDING PROTEIN Ligands: FAD, SO4 |
 |
Organism: Methanosarcina mazei, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-11-22 Classification: DNA BINDING PROTEIN Ligands: FAD, SO4 |

