Search Count: 262
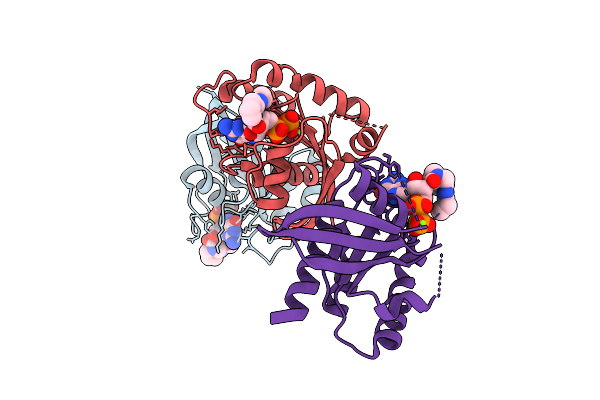 |
Crystal Structure Of Krasg13C In Complex With Nucleotide-Based Covalent Inhibitor 7B
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: A1I08, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: A1IIE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: ZN, A1IIF |
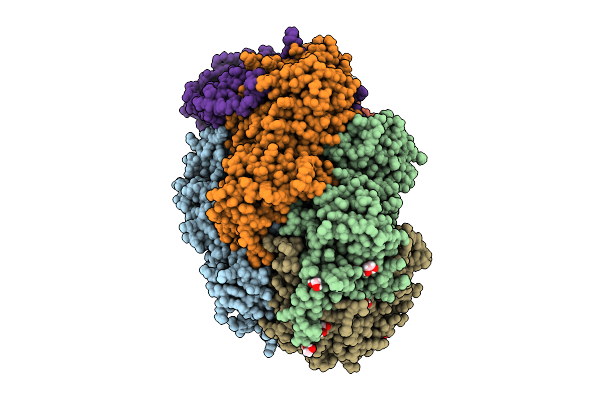 |
Methyl-Coenzyme M Reductase Of Anme-2D Candidatus Methanoperedens Vercelli Strain 1 From A Bioreactor Enrichment Culture
Organism: Candidatus methanoperedens sp.
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: TRANSFERASE Ligands: K, F43, EDO, TP7, COM, SHT |
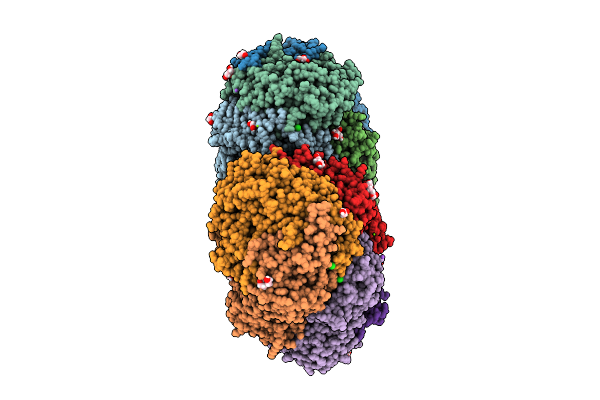 |
Krypton-Pressurized Methyl-Coenzyme M Reductase Of An Anme-2C Isolated From A Microbial Enrichment
Organism: Candidatus methanogasteraceae archaeon
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: KR, MG, PGE, EDO, CL, TP7, F43, PEG, GOL, COM, NA, K |
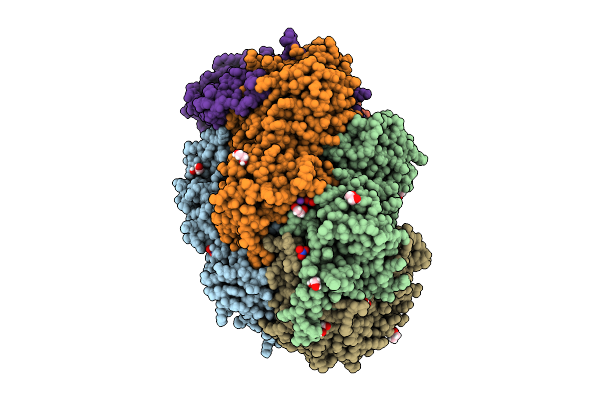 |
Methyl-Coenzyme M Reductase Of Anme-2D Candidatus Methanoperedens Sp. Blz2 From A Bioreactor Enrichment Culture
Organism: Candidatus methanoperedens sp. blz2
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE |
 |
Organism: Candidatus methanogasteraceae archaeon
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: K, NA, CL, F43, COM, TP7, GOL, MPD, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: A1IY7 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-04-09 Classification: OXIDOREDUCTASE Ligands: FE2, ZN, 8XQ, SO4, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-03-26 Classification: SUGAR BINDING PROTEIN Ligands: SCN, A1IB3 |
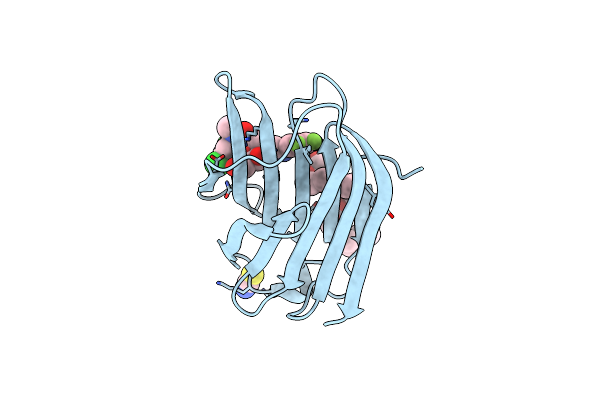 |
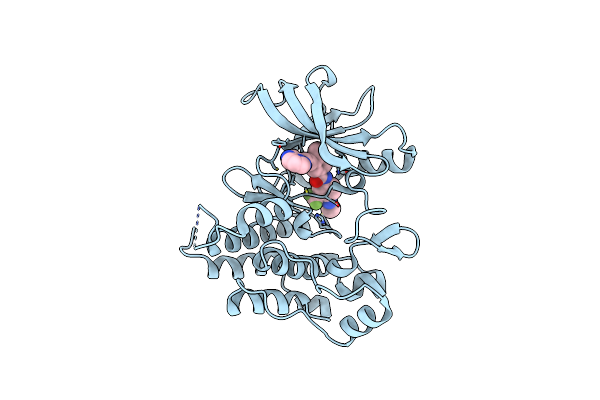
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-03-26 Classification: SUGAR BINDING PROTEIN Ligands: 2PE, A1IB4, SCN |
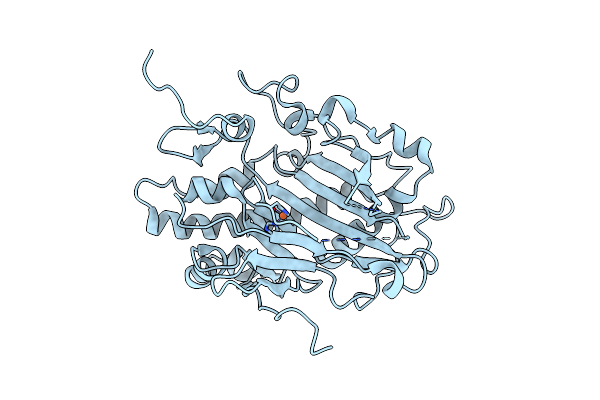 |
Crystal Structure Of Pyrimidine Nucleoside 2'-Hydroxylase (Pdn2'H) From Neurospora Crassa
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: FE |
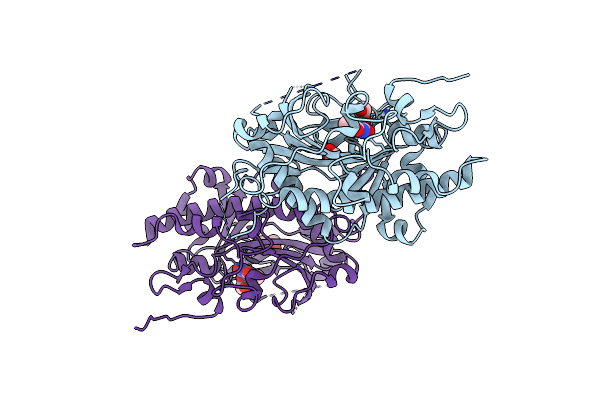 |
Crystal Structure Of Pyrimidine Nucleoside 2'-Hydroxylase (Pdn2'H) From Neurospora Crassa In Complex With Thymidine
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: MN, THM, EDO, CL, BU3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-02-12 Classification: TRANSFERASE Ligands: A1IRC |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-01-29 Classification: SUGAR BINDING PROTEIN Ligands: A1H1W, SCN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-01-29 Classification: SUGAR BINDING PROTEIN Ligands: A1H1Y, SCN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-01-29 Classification: SUGAR BINDING PROTEIN Ligands: A1H1X, SCN |
 |
Crystal Structure Of Sgvm Methyltransferase In Complex With Alpha-Ketoleucine And Zn2+ Ion
Organism: Streptomyces griseoviridis
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2024-07-24 Classification: TRANSFERASE Ligands: COI, CL, MG, ZN |
 |
Organism: Streptomyces griseoviridis
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-07-24 Classification: TRANSFERASE Ligands: ZN, COI, CL, SAM |
 |
Crystal Structure Of Alpha Keto Acid C-Methyl-Transferases Mrsa Native-Form
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: MG |

