Search Count: 17
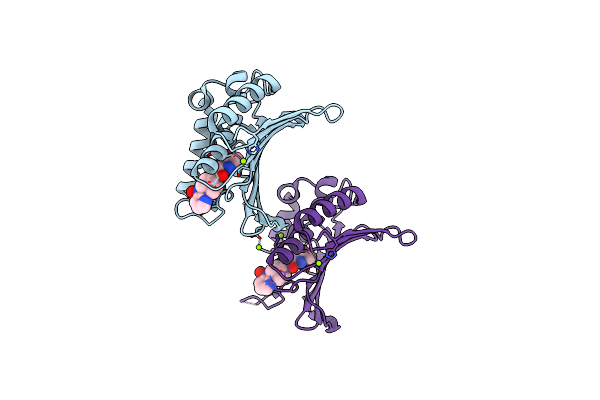 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2012-01-11 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: 08B, MG |
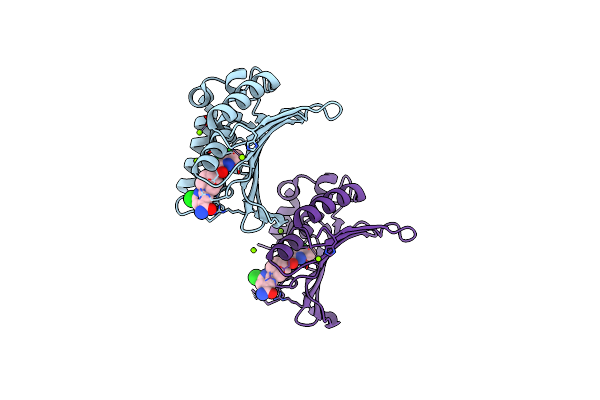 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2012-01-11 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: 087, MG |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2011-11-16 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: 07N, MG |
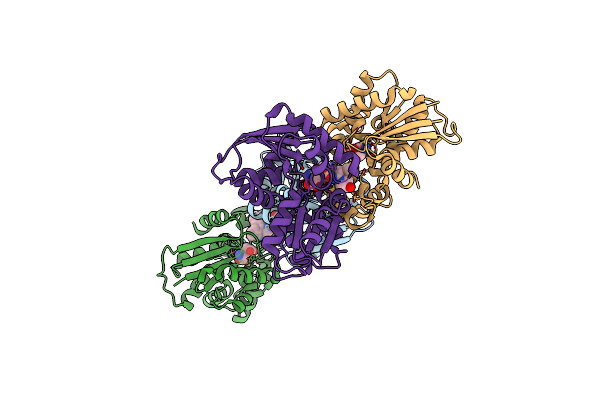 |
Crystal Structure Of Helicobacter Pylori Glutamate Racemase In Complex With D-Glutamate And An Inhibitor
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2008-12-09 Classification: ISOMERASE Ligands: DGL, VGA |
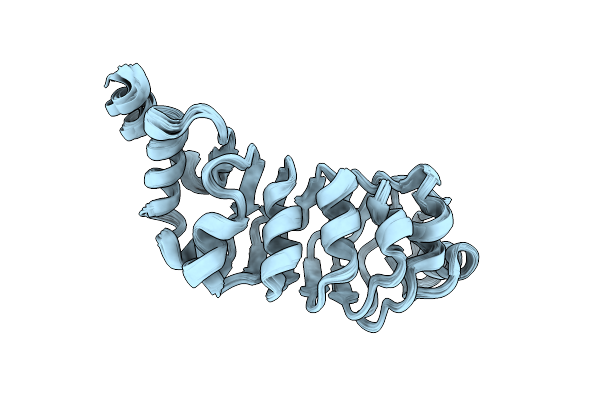 |
Relaxation-Based Refined Structure Of Chlamydomonas Outer Arm Dynein Light Chain 1
Organism: Chlamydomonas reinhardtii
Method: SOLUTION NMR Release Date: 2003-03-04 Classification: CONTRACTILE PROTEIN |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2002-01-16 Classification: DNA BINDING PROTEIN |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2001-08-01 Classification: DNA BINDING PROTEIN |
 |
 |
Organism: Chlamydomonas reinhardtii
Method: SOLUTION NMR Release Date: 2000-07-26 Classification: CONTRACTILE PROTEIN |
 |
Organism: Rattus norvegicus
Method: SOLUTION NMR Release Date: 2000-02-14 Classification: TRANSFERASE |
 |
Organism: Rattus norvegicus
Method: SOLUTION NMR Release Date: 2000-02-14 Classification: TRANSFERASE |
 |
Nmr Solution Structure Of The Single-Strand Break Repair Protein Xrcc1-N-Terminal Domain
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 1999-09-01 Classification: DNA BINDING PROTEIN |
 |
Nmr Solution Structure Of The Single-Strand Break Repair Protein Xrcc1-N-Terminal Domain
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 1999-09-01 Classification: DNA BINDING PROTEIN |
 |
Nmr Solution Structure Of The N-Terminal Domain Of Dna Polymerase Beta, Minimized Average Structure
Organism: Rattus norvegicus
Method: SOLUTION NMR Release Date: 1996-12-07 Classification: NUCLEOTIDYLTRANSFERASE (DNA-BINDING) |
 |
Nmr Solution Structure Of The N-Terminal Domain Of Dna Polymerase Beta, 55 Structures
Organism: Rattus norvegicus
Method: SOLUTION NMR Release Date: 1996-12-07 Classification: NUCLEOTIDYLTRANSFERASE (DNA-BINDING) |
 |
Determination Of The Structure Of The Dna Binding Domain Of Gamma Delta Resolvase In Solution
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 1994-11-30 Classification: SITE-SPECIFIC RECOMBINASE |
 |
Determination Of The Structure Of The Dna Binding Domain Of Gamma Delta Resolvase In Solution
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 1994-11-30 Classification: SITE-SPECIFIC RECOMBINASE |

