Search Count: 12
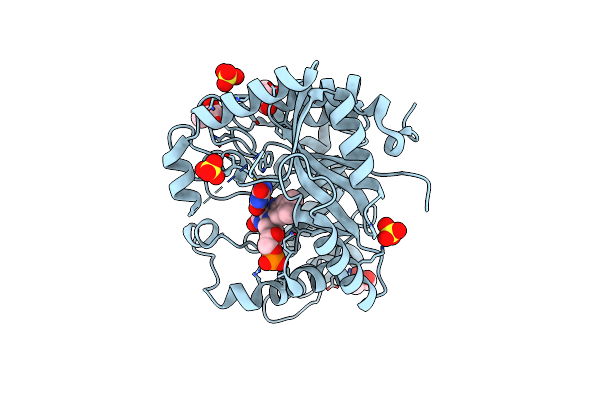 |
Crystal Structure Of A Flavoenzyme Ruta In The Pyrimidine Catabolic Pathway
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-07-19 Classification: OXIDOREDUCTASE Ligands: URA, SO4, GOL, FMN |
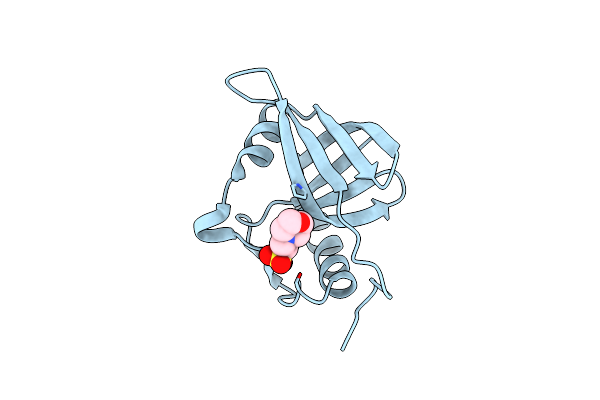 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: MES |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
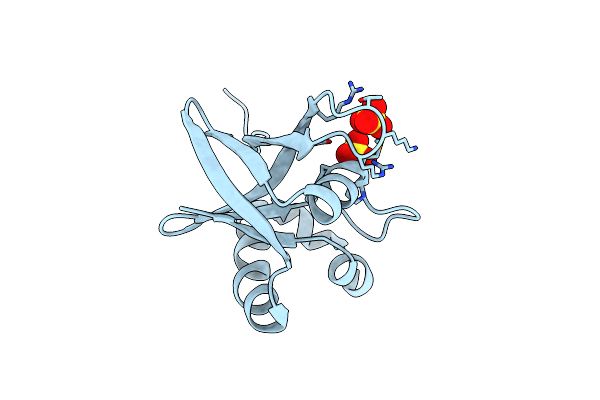
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824, With Co-Factor F420
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: F42 |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824, With Co-Factor F420
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: F42 |
 |
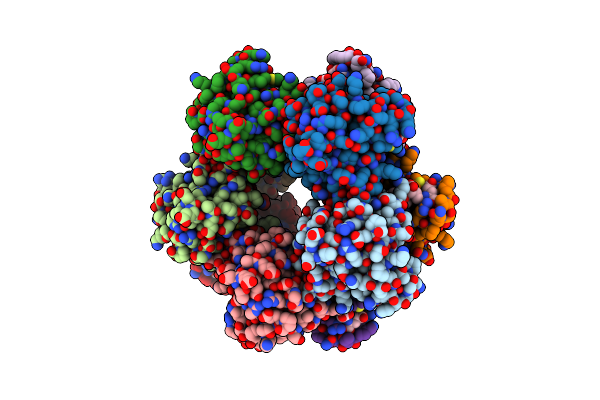
Structure Of A Deazaflavin-Dependent Nitroreductase From Nocardia Farcinica, With Co-Factor F420
Organism: Nocardia farcinica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-01-18 Classification: UNKNOWN FUNCTION Ligands: F42 |
 |
Structure Of A Deazaflavin-Dependent Reductase From Nocardia Farcinica, With Co-Factor F420
Organism: Nocardia farcinica
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-01-18 Classification: UNKNOWN FUNCTION Ligands: F42, SO4 |
 |
Organism: Mesorhizobium loti
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2010-02-09 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of 2-Methyl-3-Hydroxypyridine-5-Carboxylic Acid Oxygenase
Organism: Mesorhizobium loti
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-04-14 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Crystal Structure Of 2-Methyl-3-Hydroxypyridine-5-Carboxylic Acid Oxygenase With Substrate Bound
Organism: Mesorhizobium loti
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-04-14 Classification: OXIDOREDUCTASE Ligands: FAD, 3HM |
 |
Crystal Structure Of Mesorhizobium Loti 3-Hydroxy-2-Methylpyridine-4,5-Dicarboxylate Decarboxylase
Organism: Mesorhizobium loti
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-11-13 Classification: LYASE Ligands: MN |
 |
Crystal Structure Of Tryptophan 2,3-Dioxygenase From Ralstonia Metallidurans
Organism: Cupriavidus metallidurans
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-12-19 Classification: OXIDOREDUCTASE Ligands: HEM |

