Search Count: 198
 |
Cryo-Em Structure Of Chaetomium Thermophilum Ribosome-Bound Snd3 Translocon
Organism: Thermochaetoides thermophila dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RIBOSOME Ligands: MG |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSPORT PROTEIN Ligands: CLR, SD9 |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSPORT PROTEIN Ligands: WRF, CLR |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSPORT PROTEIN Ligands: CLR, MF8 |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSPORT PROTEIN Ligands: CLR |
 |
Y100F Mutant Of E. Coli Dihydrofolate Reductase Complexed With Nadp+ (Oxidized Form)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: OXIDOREDUCTASE Ligands: NAP, CA |
 |
Crystal Structure Of N-Terminal Domain Of Borealin (20-88) In Complex With Synthetic Antibody Fragment
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: IMMUNE SYSTEM Ligands: MPD, CL |
 |
Horse Liver Alcohol Dehydrogenase F93W In Complex With Nadh And N-Cylcohexyl Formamide
Organism: Equus caballus
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, CXF |
 |
Horse Liver Alcohol Dehydrogenase V203L In Complex With Nadh And N-Cylcohexyl Formamide
Organism: Equus caballus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, CXF |
 |
Horse Liver Alcohol Dehydrogenase V203A In Complex With Nadh And N-Cylcohexyl Formamide
Organism: Equus caballus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, CXF |
 |
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-06-04 Classification: APOPTOSIS |
 |
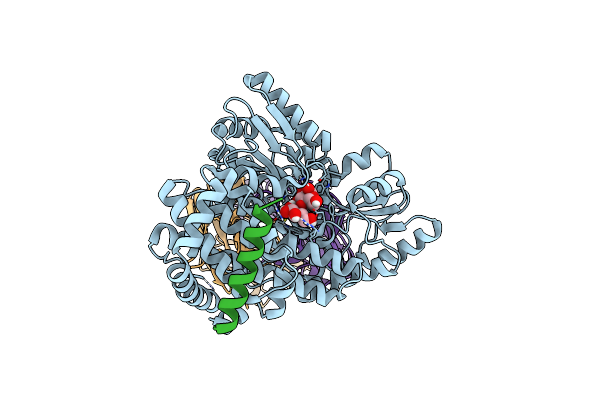
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-06-04 Classification: APOPTOSIS |
 |
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: APOPTOSIS |
 |
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-06-04 Classification: APOPTOSIS |
 |
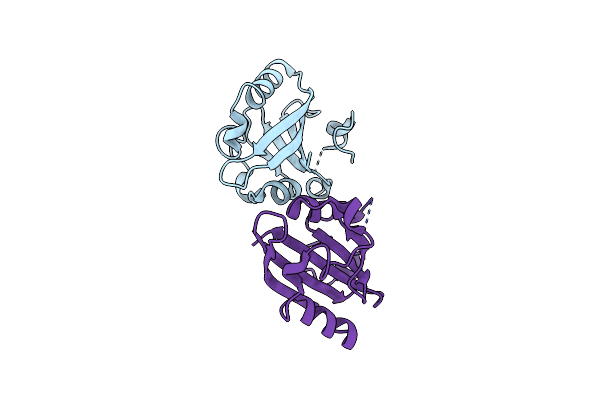
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: MN |
 |
Organism: Halorhodospira halophila
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: CL |
 |
L54A Mutant Of E. Coli Dihydrofolate Reductase Complexed With Nicotinamide Adenine Dinucleotide Phosphate (Oxidized Form)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2025-04-23 Classification: OXIDOREDUCTASE Ligands: NAP, CA |
 |
Organism: Bos taurus, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: CLR |
 |
Organism: Severe acute respiratory syndrome coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: PROTEIN BINDING Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: PROTEIN BINDING |

