Search Count: 21
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-11-05 Classification: ISOMERASE Ligands: 9CK, EDO, 1PE, PGE, PO4, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2025-11-05 Classification: ISOMERASE Ligands: A1IU3 |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME |
 |
Organism: Escherichia coli k-12, Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME |
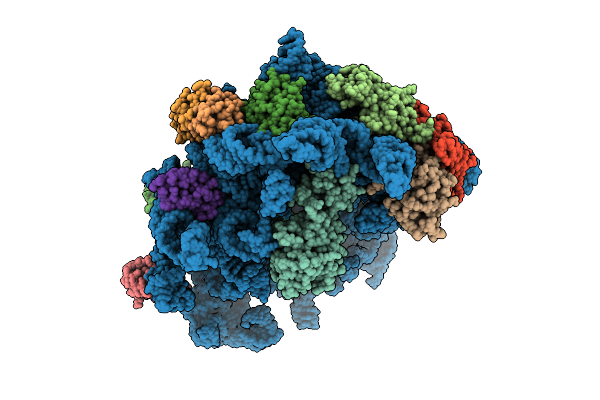 |
Organism: Escherichia coli k-12, Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME Ligands: AN6 |
 |
Organism: Legionella pneumophila str. lens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TOXIN |
 |
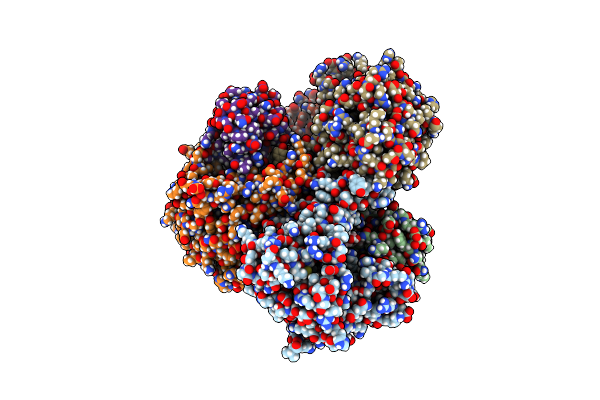
Organism: Legionella pneumophila
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TOXIN |
 |
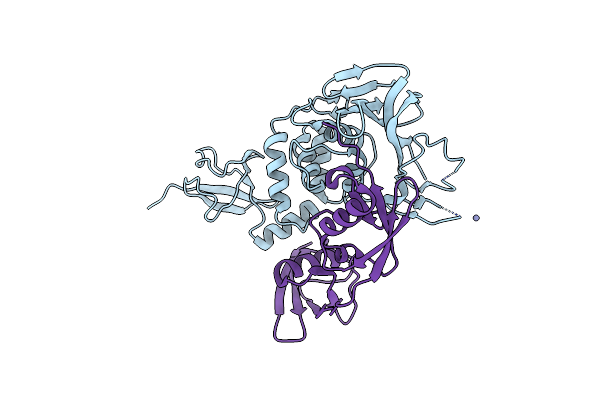
Organism: Legionella pneumophila subsp. pneumophila, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2024-10-16 Classification: UNKNOWN FUNCTION Ligands: A1H50 |
 |
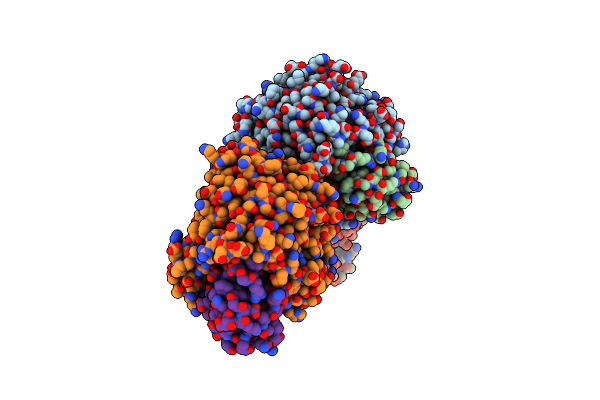
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2020-05-13 Classification: CELL INVASION Ligands: ZN |
 |
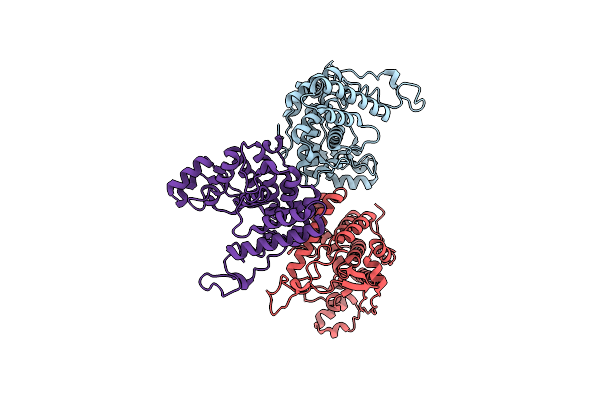
Organism: Legionella pneumophila subsp. pneumophila, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2019-11-13 Classification: CELL INVASION |
 |
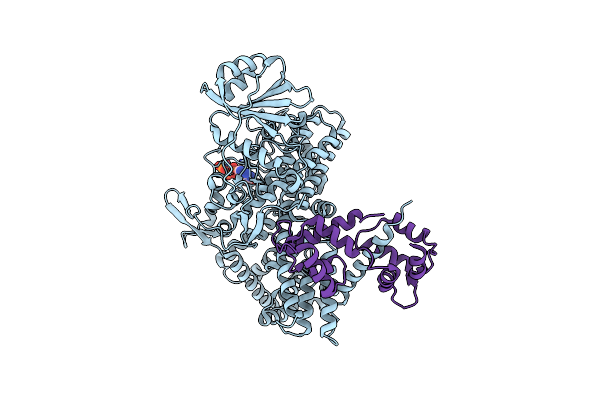
Organism: Legionella pneumophila subsp. pneumophila
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2019-11-13 Classification: CELL INVASION |
 |
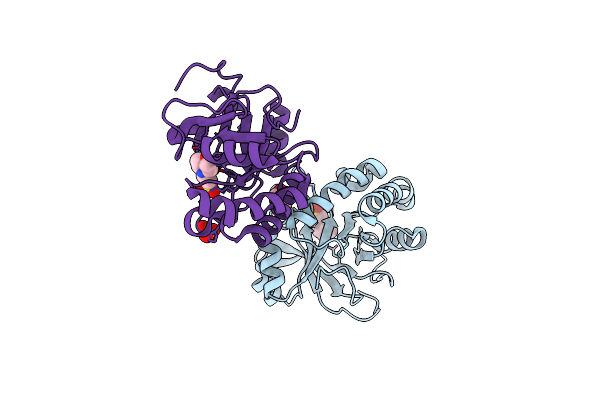
Organism: Legionella pneumophila, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: TRANSFERASE Ligands: ANP |
 |
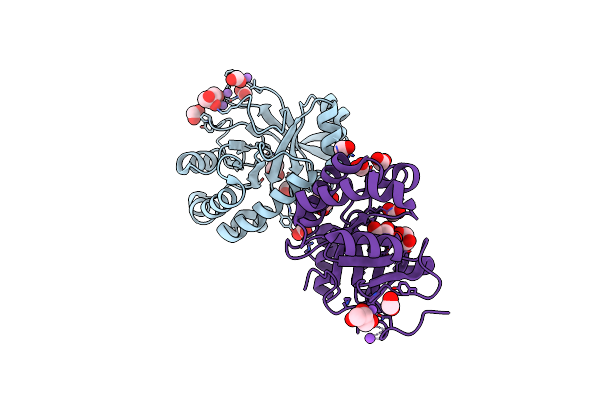
Crystal Structure Of The Mycobacterium Tuberculosis Tat-Secreted Protein Rv2525C In Complex With Mes (Monoclinic Crystal Form I)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2014-10-08 Classification: UNKNOWN FUNCTION Ligands: MES, GOL, SO4 |
 |
Crystal Structure Of The Mycobacterium Tuberculosis Tat-Secreted Protein Rv2525C, Monoclinic Crystal Form I
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2014-10-08 Classification: UNKNOWN FUNCTION Ligands: GOL, FMT, NA |
 |
Crystal Structure Of The Mycobacterium Tuberculosis Tat-Secreted Protein Rv2525C In Complex With L-Tartrate (Orthorhombic Crystal Form)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2014-10-08 Classification: UNKNOWN FUNCTION Ligands: TLA, GOL |
 |
Crystal Structure Of The Mycobacterium Tuberculosis Tat-Secreted Protein Rv2525C In Complex With Hepes (Monoclinic Crystal Form Ii)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2014-10-08 Classification: UNKNOWN FUNCTION Ligands: EPE, NA |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2012-10-03 Classification: OXIDOREDUCTASE Ligands: NAI |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-10-03 Classification: OXIDOREDUCTASE Ligands: NAI |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-10-03 Classification: OXIDOREDUCTASE Ligands: NAI |
 |
Crystal Structure Of Peroxisome Proliferator-Activatedeceptor Alpha (Pparalpha) Complex With N-3-((2-(4-Chlorophenyl)-5-Methyl-1,3-Oxazol-4-Yl)Methoxy)Benzyl)-N-(Methoxycarbonyl)Glycine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-04-28 Classification: HORMONE RECEPTOR Ligands: 7HA |

