Search Count: 21
 |
Organism: Marine metagenome
Method: X-RAY DIFFRACTION Release Date: 2024-10-09 Classification: SPLICING Ligands: FMT, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-05-27 Classification: SPLICING Ligands: GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2020-05-27 Classification: SPLICING Ligands: SO4, GOL |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2019-11-27 Classification: NUCLEAR PROTEIN |
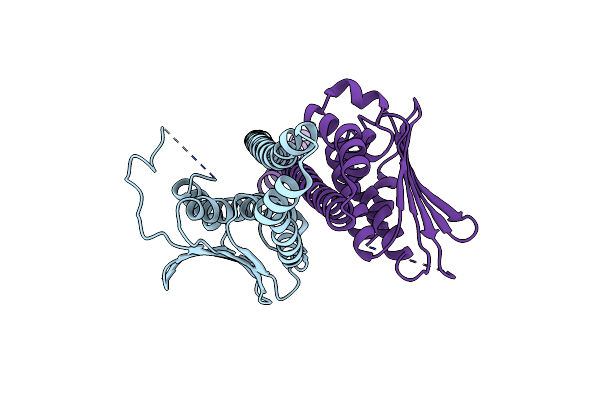 |
Chimeric Structure Of Saccharomyces Cerevisiae Gcn4 Leucine Zipper Fused To Staphylococcus Aureus Agrc Cytoplasmic Histidine Kinase Module (Dataset Anisotropically Truncated By Staraniso)
Organism: Saccharomyces cerevisiae, Staphylococcus aureus subsp. aureus z172
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2019-02-27 Classification: SIGNALING PROTEIN |
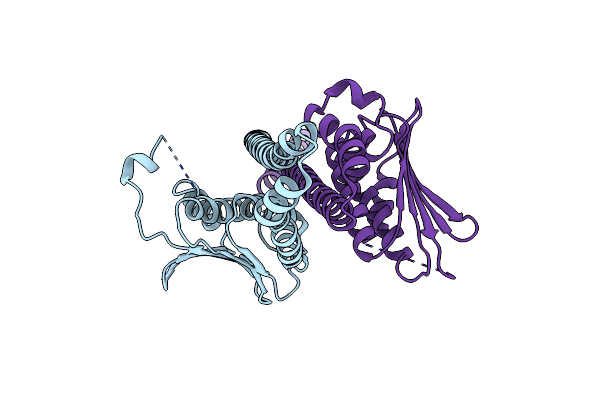 |
Chimeric Structure Of Saccharomyces Cerevisiae Gcn4 Leucine Zipper Fused To Staphylococcus Aureus Agrc Cytoplasmic Histidine Kinase Module (Dataset Isotropically Truncated By Hkl2000)
Organism: Saccharomyces cerevisiae, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-02-27 Classification: SIGNALING PROTEIN |
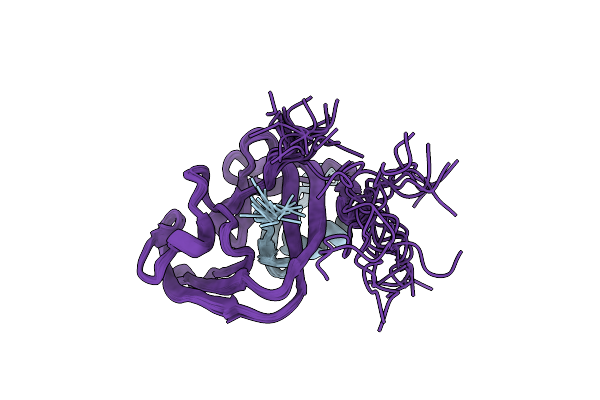 |
Organism: Enterobacteria phage t7
Method: SOLUTION NMR Release Date: 2018-09-19 Classification: SPLICING |
 |
Organism: Mycobacterium xenopi
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2014-05-14 Classification: ISOMERASE Ligands: MG |
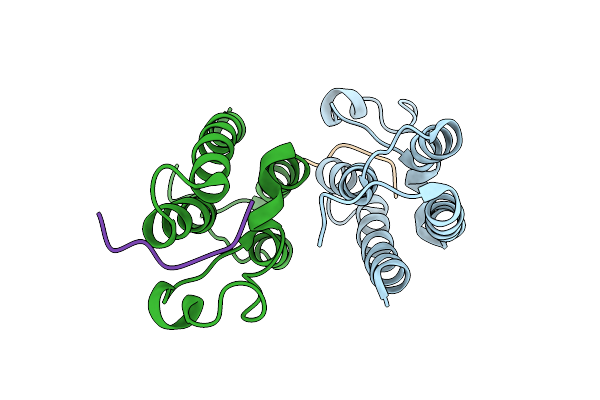 |
Organism: Homo sapiens, Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-06-01 Classification: TRANSCRIPTION/NUCLEAR PROTEIN |
 |
Organism: Homo sapiens, Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-06-01 Classification: TRANSCRIPTION/NUCLEAR PROTEIN Ligands: GOL |
 |
Crystal Structure Of Bptf Phd-Linker-Bromo In Complex With Histone H4K12Ac Peptide
Organism: Homo sapiens, Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-06-01 Classification: TRANSCRIPTION/NUCLEAR PROTEIN Ligands: ZN |
 |
Autoregulation Of A Group 1 Bacterial Sigma Factor Involves The Formation Of A Region 1.1- Induced Compacted Structure
Organism: Thermotoga maritima
Method: SOLUTION NMR Release Date: 2008-10-28 Classification: TRANSCRIPTION |
 |
Ion Selectivity In A Semi-Synthetic K+ Channel Locked In The Conductive Conformation
Organism: Streptomyces lividans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-11-21 Classification: MEMBRANE PROTEIN Ligands: K, 1EM |
 |
Ion Selectivity In A Semi-Synthetic K+ Channel Locked In The Conductive Conformation
Organism: Streptomyces lividans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2006-11-21 Classification: MEMBRANE PROTEIN Ligands: K, 1EM |
 |
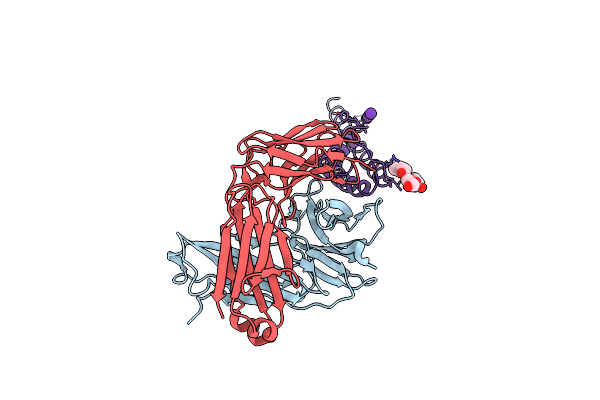
Structure Of A K Channel With An Amide To Ester Substitution In The Selectivity Filter
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2006-09-12 Classification: MEMBRANE PROTEIN Ligands: K, B3H |
 |
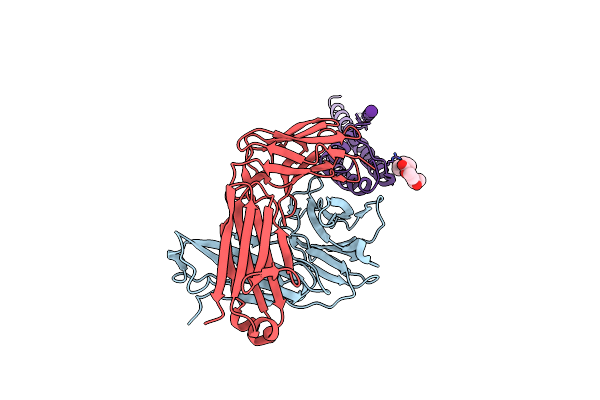
Rb+ Complex Of A K Channel With An Amide To Ester Substitution In The Selectivity Filter
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2006-09-12 Classification: MEMBRANE PROTEIN Ligands: RB, B3H |
 |
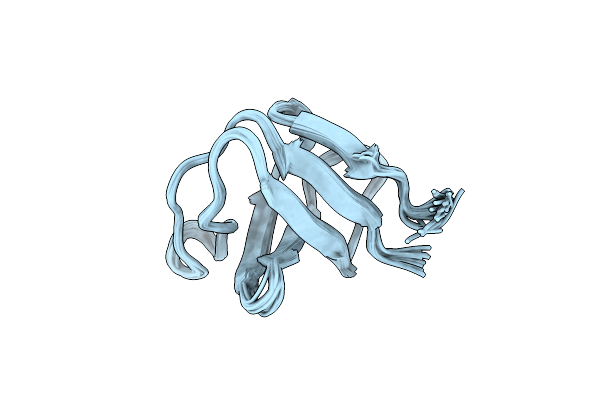
Cs+ Complex Of A K Channel With An Amide To Ester Substitution In The Selectivity Filter
Organism: Synthetic construct, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2006-09-12 Classification: MEMBRANE PROTEIN Ligands: CS, B3H |
 |
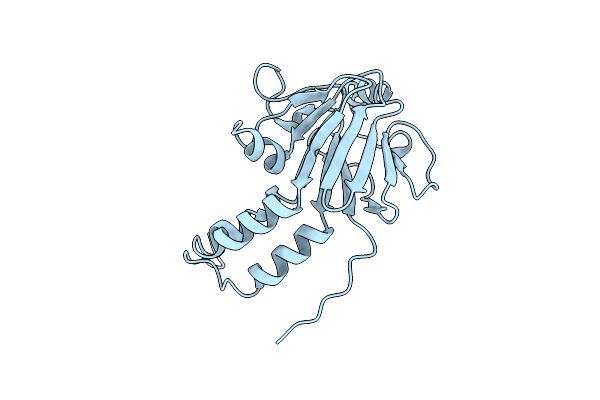
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2006-08-01 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2002-02-06 Classification: TRANSCRIPTION |
 |
Cytoplasmic Domain Of Unphosphorylated Type I Tgf-Beta Receptor Crystallized Without Fkbp12
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2001-10-03 Classification: TRANSFERASE Ligands: SO4 |

