Search Count: 27
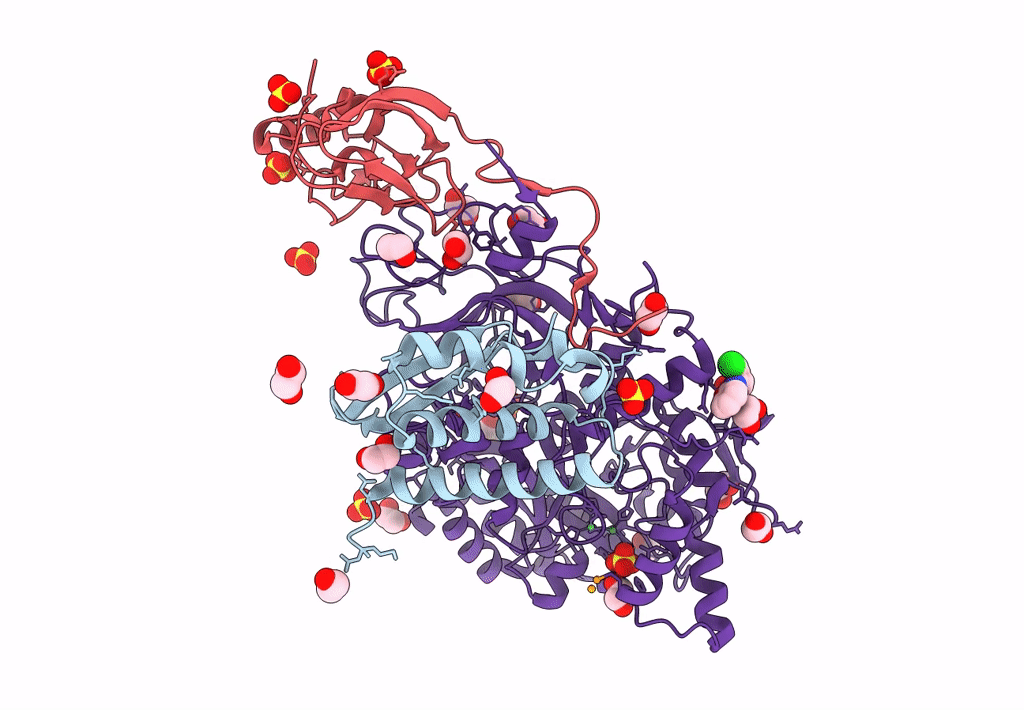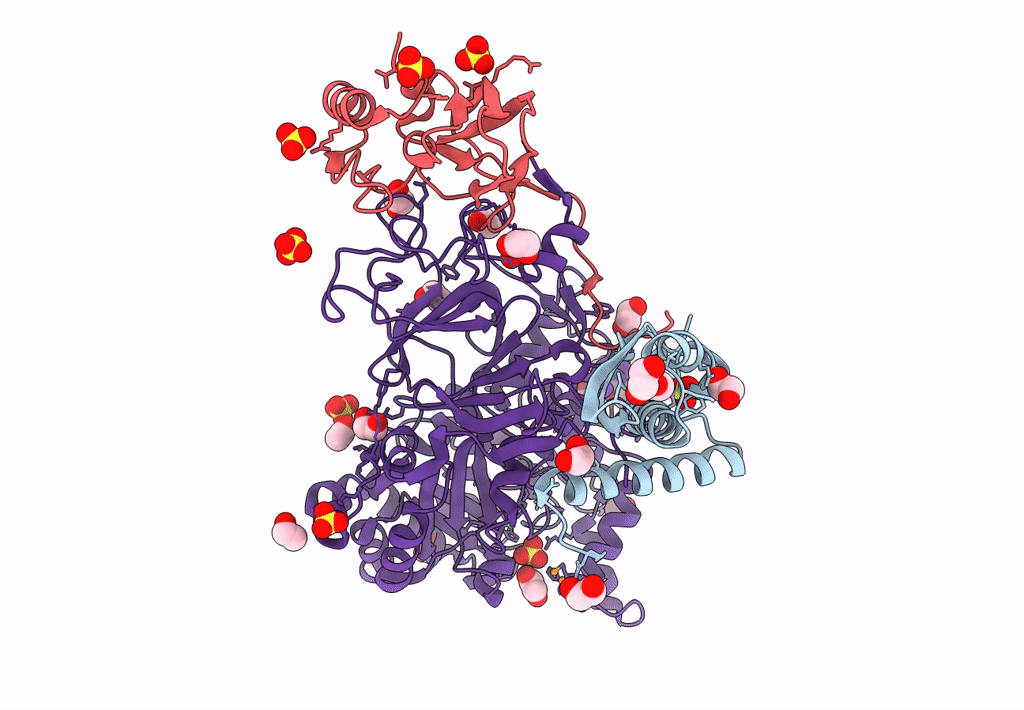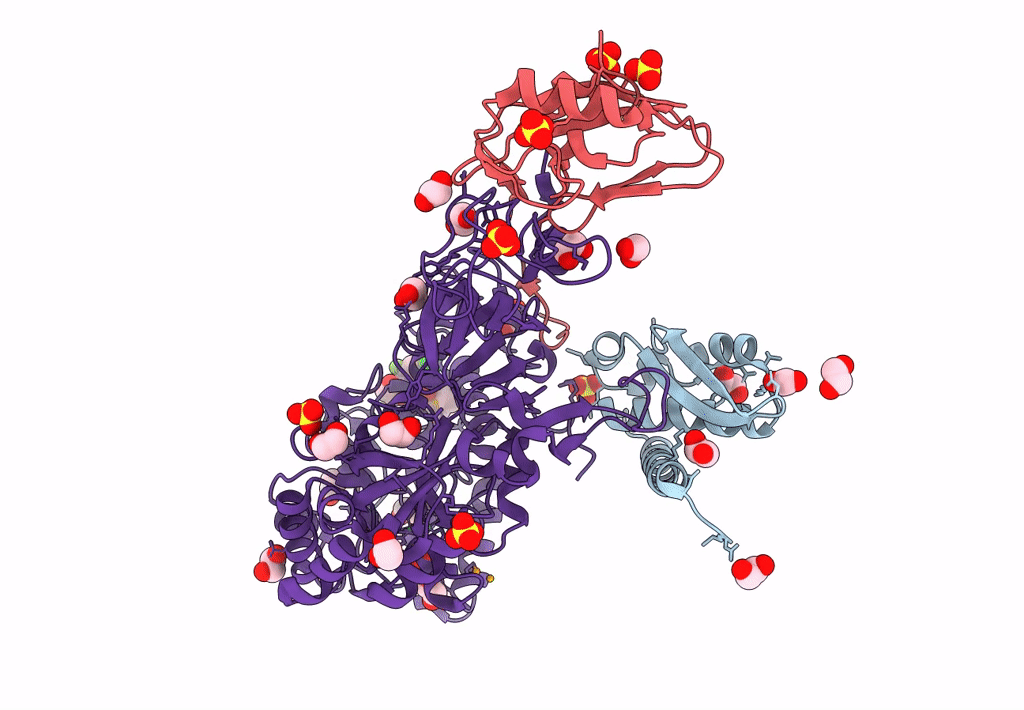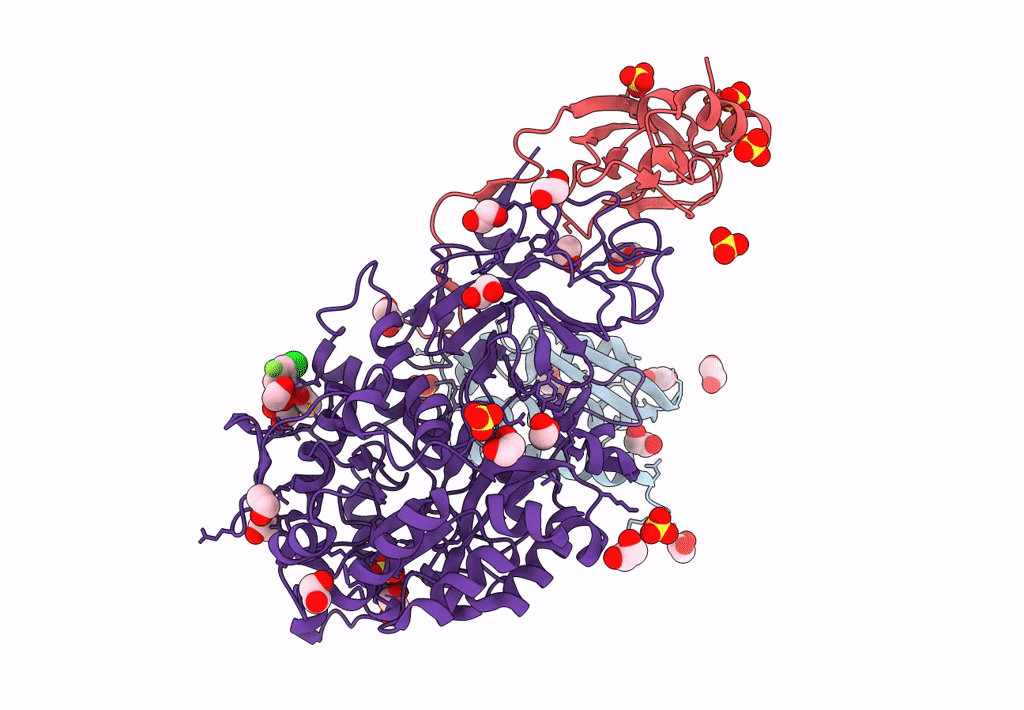 |
Sporosarcina Pasteurii Urease (Spu) Co-Crystallized In The Presence Of An Ebselen-Derivative And Bound To Se Atoms
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: EDO, SO4, NI, OH, IU9, SE |
 |
Crystal Structure Of Ppk2 Class Iii In Complex With Adp From Cytophaga Hutchinsonii Atcc 33406
Organism: Cytophaga hutchinsonii (strain atcc 33406 / ncimb 9469)
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: ADP, GOL |
 |
Crystal Structure Of Ppk2 Class Iii In The Complex With Amp From Cytophaga Hutchinsonii Atcc 33406
Organism: Cytophaga hutchinsonii (strain atcc 33406 / ncimb 9469)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: AMP, CL |
 |
Crystal Structure Of Ppk2 Class Iii In Complex With Guanosine 5-Tetraphosphate
Organism: Cytophaga hutchinsonii (strain atcc 33406 / ncimb 9469)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: BKP |
 |
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: ATP, MG, GOL, MPD, CL |
 |
Organism: Cytophaga hutchinsonii (strain atcc 33406 / ncimb 9469)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-01-16 Classification: transferase/transferase inhibitor Ligands: BOY, GOL, SRT |
 |
Crystal Structure Of Ppk2 (Class Iii) In Complex With Bisphosphonate Inhibitor (2-((3,5-Dichlorophenyl)Amino)Ethane-1,1-Diyl)Diphosphonic Acid
Organism: Cytophaga hutchinsonii (strain atcc 33406 / ncimb 9469)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-01-16 Classification: transferase/transferase inhibitor Ligands: BWJ, GOL |
 |
Organism: Cytophaga hutchinsonii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-01-16 Classification: transferase/transferase inhibitor Ligands: C8A, PO4, GOL |
 |
The Crystal Structure Of Aminopeptidase N In Complex With N-Benzyl-1,2-Diaminoethylphosphonic Acid
Organism: Neisseria meningitidis serogroup b (strain mc58)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-11-25 Classification: hydrolase/hydrolase inhibitor Ligands: ZN, SO4, 5HR, GOL, IMD |
 |
Crystal Structure Of Aminopeptidase N In Complex With The Phosphinic Dipeptide Analogue Ll-(R,S)-Hphep[Ch2]Phe
Organism: Neisseria meningitidis mc58
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-10-01 Classification: HYDROLASE Ligands: 37B, ZN, GOL, IMD, SO4 |
 |
Crystal Structure Of Aminopeptidase N In Complex With The Phosphinic Dipeptide Analogue Ll-(R,S)-Hphep[Ch2]Phe(4-Ch2Nh2)
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-09-24 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 32Q, 32R, IMD, GOL, ZN, SO4 |
 |
Crystal Structure Of Aminopeptidase N In Complex With The Phosphinic Dipeptide Analogue Ll-(R,S)-2-(Pyridin-3-Yl)Ethylglyp[Ch2]Phe
Organism: Neisseria meningitidis mc58
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-09-24 Classification: HYDROLASE Ligands: 379, GOL, IMD, ZN, SO4 |
 |
Crystal Structure Of Aminopeptidase N In Complex With N-Cyclohexyl-1,2-Diaminoethylphosphonic Acid
Organism: Neisseria meningitidis mc58
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-09-24 Classification: HYDROLASE Ligands: 37E, ZN, SO4 |
 |
Crystal Structure Of Aminopeptidase N In Complex With The Phosphinic Dipeptide Analogue Ll-(R,S)-Hphep[Ch2]Phe(3-Ch2Nh2)
Organism: Neisseria meningitidis mc58
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-09-10 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 3DZ, GOL, IMD, ZN, SO4 |
 |
Crystal Structure Of Aminopeptidase N In Complex With The Phosphonic Acid Analogue Of Leucine L-(R)-Leup
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: PLU, GOL, ZN, SO4 |
 |
Crystal Structure Of Aminopeptidase N In Complex With The Phosphonic Acid Analogue Of Leucine (D-(S)-Leup)
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: 2WW, ZN, SO4, PO4 |
 |
Crystal Structure Of Aminopeptidase N In Complex With Phosphonic Acid Analogue Of Homophenylalanine L-(R)-Hphep
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-06-25 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2X0, ZN, IMD, GOL, SO4 |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2010-02-02 Classification: HYDROLASE Ligands: ZN, CO3, SO4, 1PE, 2PE |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2010-02-02 Classification: HYDROLASE Ligands: CO3, ZN, SO4, 1PE, 2PE |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-02-02 Classification: HYDROLASE Ligands: CO3, ZN, BES, MG, SO4, 1PE |

