Search Count: 36
 |
Crystal Structure Of Hydrogen Sulfide-Bound Superoxide Dismutase In Oxidized State
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-09-06 Classification: OXIDOREDUCTASE Ligands: CU, ZN, EDO, GOL, SO4, CL, H2S |
 |
Crystal Structure Of Hydrogen Sulfide-Bound Superoxide Dismutase In Reduced State
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-09-06 Classification: OXIDOREDUCTASE Ligands: CU, ZN, EDO, H2S, SO4, CL, GOL |
 |
Organism: Danio rerio, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: LIPID TRANSPORT Ligands: ZGS, LMT, NA |
 |
Organism: Danio rerio, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: LIPID TRANSPORT Ligands: LMT |
 |
Organism: Danio rerio, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: LIPID TRANSPORT Ligands: ZGS, LMT, NA |
 |
Organism: Danio rerio, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: LIPID TRANSPORT Ligands: ZGS, LMT, NA |
 |
Organism: Danio rerio, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: LIPID TRANSPORT Ligands: ZGS, LMT |
 |
Organism: Danio rerio, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: LIPID TRANSPORT Ligands: ZGS, LMT |
 |
Organism: Exiguobacterium sibiricum (strain dsm 17290 / cip 109462 / jcm 13490 / 255-15)
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2023-04-26 Classification: OXIDOREDUCTASE Ligands: GOL, CA |
 |
The Crystal Structure Of Vypal2-C214A, A Dead Mutant Of Vypal2 Peptide Asparaginyl Ligase In Form Ii
Organism: Viola philippica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-07-13 Classification: PLANT PROTEIN Ligands: NAG |
 |
The Crystal Structure Of Vypal2-I244V, A More Efficient Mutant Of Vypal2 Peptide Asparaginyl Ligase In Its Active Enzyme Form
Organism: Viola philippica
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2022-06-29 Classification: PLANT PROTEIN Ligands: NAG, EDO, GOL |
 |
The Crystal Structure Of Vypal2-C214A, A Dead Mutant Of Vypal2 Peptide Asparaginyl Ligase In Form I
Organism: Viola philippica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-06-29 Classification: PLANT PROTEIN |
 |
The Crystal Structure Of Vypal2 Peptide Asparaginyl Ligase In Its Active Enzyme Form
Organism: Viola philippica
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-06-29 Classification: PLANT PROTEIN Ligands: NAG, EDO |
 |
Cryo-Em Structure Of Prenyltransferase Domain Of Macrophoma Phaseolina Macrophomene Synthase
Organism: Macrophomina phaseolina ms6
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: TRANSFERASE |
 |
Organism: Talaromyces verruculosus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-06-08 Classification: TRANSFERASE |
 |
Organism: Talaromyces verruculosus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-06-08 Classification: TRANSFERASE Ligands: NI |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2021-01-13 Classification: ANTITOXIN |
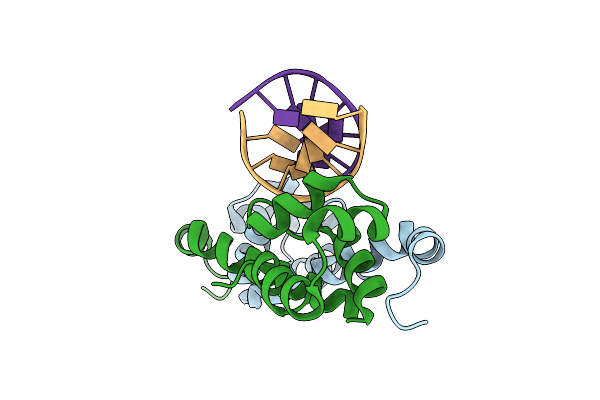 |
Organism: Pseudomonas aeruginosa pao1, Pseudomonas aeruginosa ucbpp-pa14
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-01-13 Classification: ANTITOXIN/DNA |
 |
Organism: Pseudomonas aeruginosa pao1, Pseudomonas aeruginosa ucbpp-pa14
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2021-01-13 Classification: ANTITOXIN/DNA Ligands: TMP |
 |
Crystal Structure Of Sodium-Coupled Neutral Amino Acid Transporter Slc38A9 In The N-Terminal Plugged Form
Organism: Danio rerio, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2020-12-23 Classification: TRANSPORT PROTEIN |

