Search Count: 17
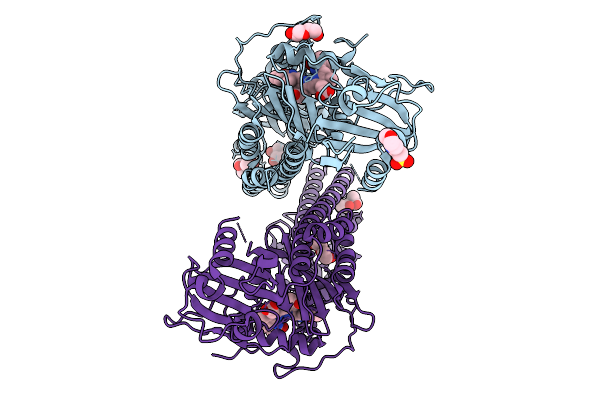 |
Crystal Structure Of The Photosensory Core Module (Pcm) Of A Cyano-Phenylalanine Mutant Ocnf205 Of The Bathy Phytochrome Agp2 From Agrobacterium Fabrum In The Pfr State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, MPD, PEG, MES, CL |
 |
Organism: Synechocystis
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2025-03-12 Classification: FLUORESCENT PROTEIN Ligands: CYC |
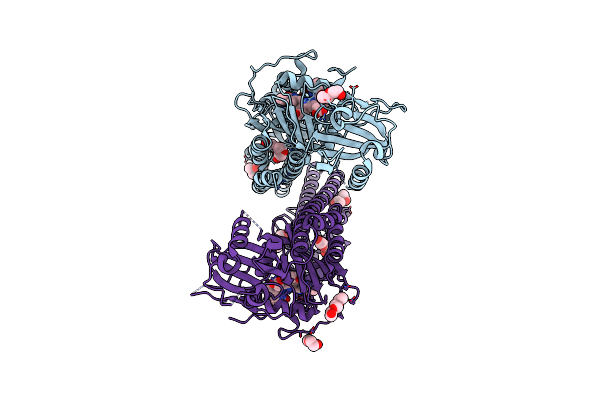 |
Crystal Structure Of The Photosensory Core Module (Pcm) Of A Cyano-Phenylalanine Mutant Ocnf165 Of The Bathy Phytochrome Agp2 From Agrobacterium Fabrum In The Pfr State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-11-27 Classification: SIGNALING PROTEIN Ligands: P6G, MPD, PEG, EL5, CL |
 |
Crystal Structure Of The Photosensory Core Module (Pcm) Of A Cyano-Phenylalanine Mutant Ocnf192 Of The Bathy Phytochrome Agp2 From Agrobacterium Fabrum In The Pfr State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2024-11-27 Classification: SIGNALING PROTEIN Ligands: PEG, MPD, EL5, CL |
 |
Crystal Structure Of The Dark-Adapted Full-Length Bacteriophytochrome Xccbphp-G454E Variant From Xanthomonas Campestris In The Pfr State
Organism: Xanthomonas campestris pv. campestris (strain 8004)
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2021-12-29 Classification: SIGNALING PROTEIN Ligands: BLA |
 |
Crystal Structure Of The Photosensory Module From Xanthomonas Campestris Bacteriophytochrome Xccbphp In The Pfr State
Organism: Xanthomonas campestris pv. campestris (strain 8004)
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2021-12-29 Classification: SIGNALING PROTEIN Ligands: BLA |
 |
Crystal Structure Of The Dark-Adapted Full-Length Bacteriophytochrome Xccbphp From Xanthomonas Campestris In The Pr State Bound To Bv Chromophore
Organism: Xanthomonas campestris pv. campestris (strain 8004)
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2020-12-30 Classification: SIGNALING PROTEIN Ligands: BLA |
 |
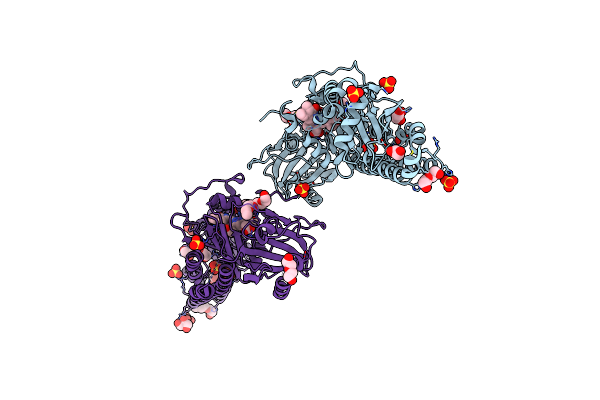
Crystal Structure Of The Photosensory Core Module (Pcm) Of A Bathy Phytochrome From Agrobacterium Fabrum In The Pfr State.
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-11-28 Classification: SIGNALING PROTEIN Ligands: EL5 |
 |
Crystal Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State.
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2018-11-28 Classification: FLUORESCENT PROTEIN Ligands: EL5, SO4, GOL, PEG, ETE, PG0 |
 |
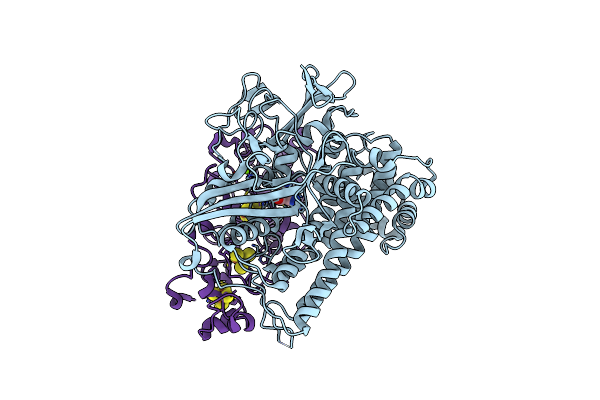
Crystal Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Functional Meta-F Intermediate State.
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2018-11-28 Classification: FLUORESCENT PROTEIN Ligands: EL5, SO4, CL, GOL, PEG, ETE, PG0 |
 |
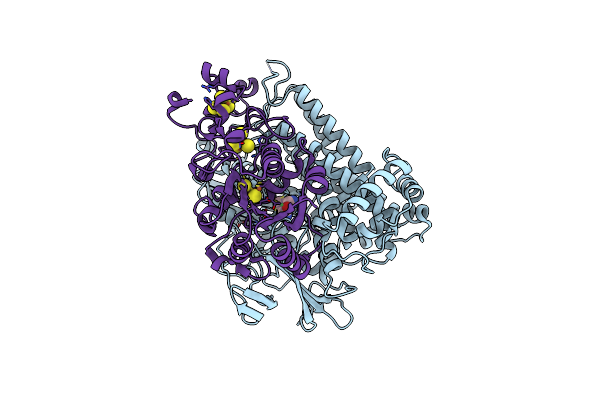
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In A Its As-Isolated High-Pressurized Form
Organism: Ralstonia eutropha h16
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2018-02-21 Classification: OXIDOREDUCTASE Ligands: NFV, MG, SF4, F3S, F4S, CL |
 |
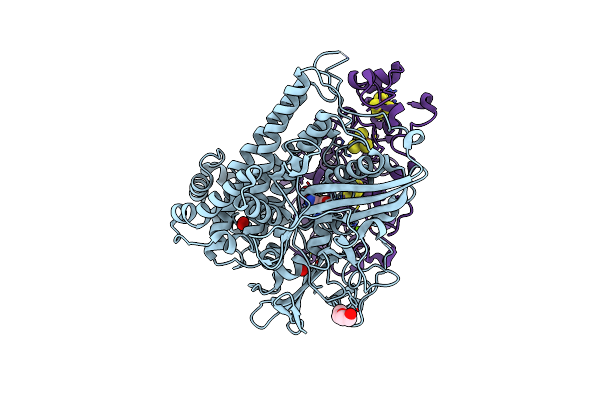
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In Its As-Isolated Form (Oxidized State - State 3)
Organism: Ralstonia eutropha h16
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-02-21 Classification: OXIDOREDUCTASE Ligands: MG, CL, NFV, SF4, F3S, F4S |
 |
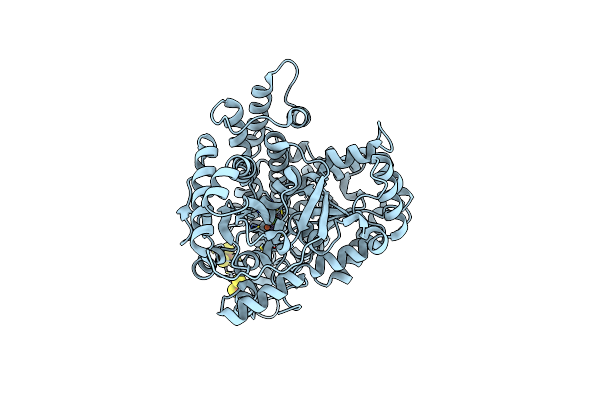
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In Its O2-Derivatized Form By A "Soak-And-Freeze" Derivatization Method
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2018-02-21 Classification: OXIDOREDUCTASE Ligands: NFV, MG, OXY, PEG, SF4, F3S, F4S, CL, O |
 |
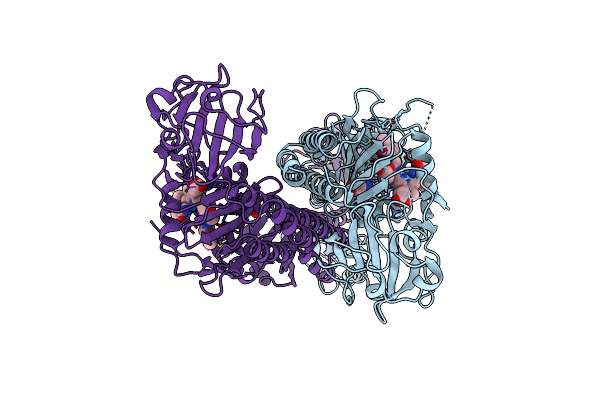
Organism: Carboxydothermus hydrogenoformans
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2016-09-14 Classification: OXIDOREDUCTASE Ligands: SF4, FES, 82N, FE2 |
 |
Crystal Structure Of The Dark-Adapted Full-Length Bacteriophytochrome Xccbphp From Xanthomonas Campestris Bound To Bv Chromophore
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2016-05-04 Classification: SIGNALING PROTEIN Ligands: BLA, TRS, CL |
 |
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In Its As-Isolated Form - Oxidized State 3
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2015-01-28 Classification: OXIDOREDUCTASE Ligands: NFV, PO4, MG, CL, NA, SF4, F3S, F4S |
 |
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-10-30 Classification: TRANSFERASE Ligands: GLU, CYC, GOL, FMT |

