Search Count: 118
 |
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-04-07 Classification: TRANSFERASE Ligands: GOL, EDO |
 |
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2011-07-20 Classification: Transcription regulator |
 |
Crystal Structure Of Conserved Protein Of Unknown Function Ef_1977 From Enterococcus Faecalis
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2010-07-28 Classification: Structural Genomics, Unknown function Ligands: ZN, CIT |
 |
A Putative C-Terminal Regulatory Domain Of Aspartate Kinase From Porphyromonas Gingivalis W83.
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2010-04-14 Classification: TRANSFERASE Ligands: SO4 |
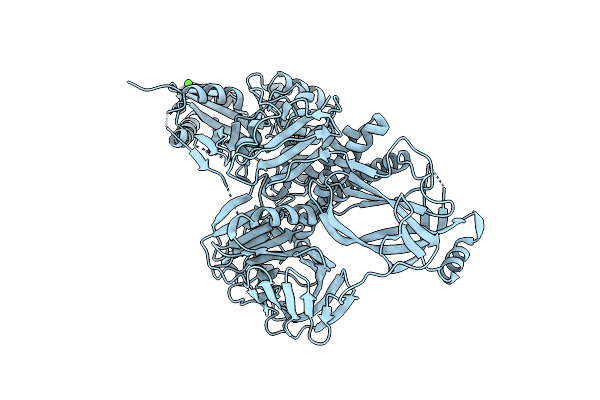 |
The Crystal Structure Of A Protein In The Glycosyl Hydrolase Family 38 From Enterococcus Faecalis To 2.55A
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2010-03-16 Classification: HYDROLASE Ligands: CA |
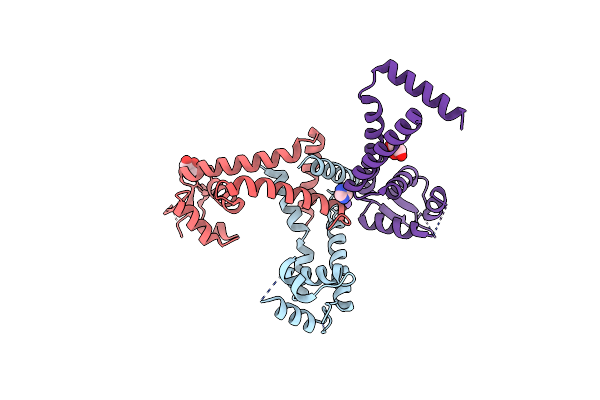 |
The Structure Of A Putative Marr Family Transcriptional Regulator From Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-12-22 Classification: Transcription regulator Ligands: K, GOL, IMD |
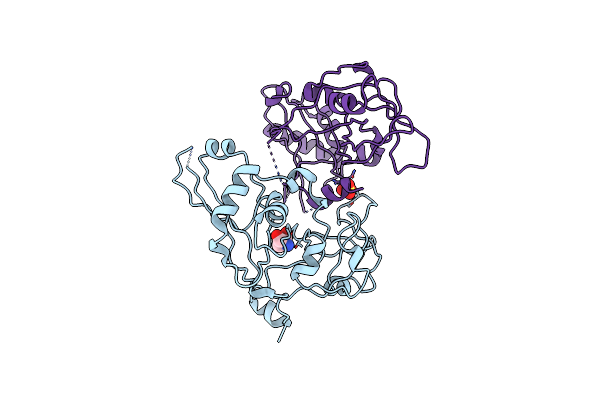 |
The Crystal Structure Of A Domain Of A Putative Permease Protein From Porphyromonas Gingivalis To 2A
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-09-08 Classification: TRANSPORT PROTEIN Ligands: TRS, SO4 |
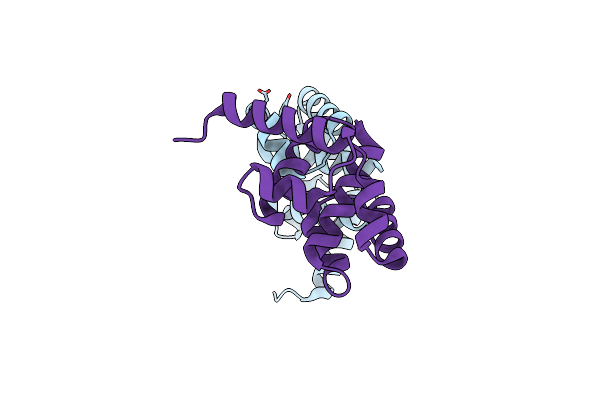 |
The Crystal Structure Of The C-Terminal Domain Of The Atp-Dependent Dna Helicase Recq From Porphyromonas Gingivalis To 1.6A
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-09-08 Classification: HYDROLASE |
 |
The Structure Of A Putative Transcriptional Regulator Tetr Family Protein From Vibrio Parahaemolyticus.
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-07-07 Classification: transcription regulator Ligands: SO4, GOL, CL |
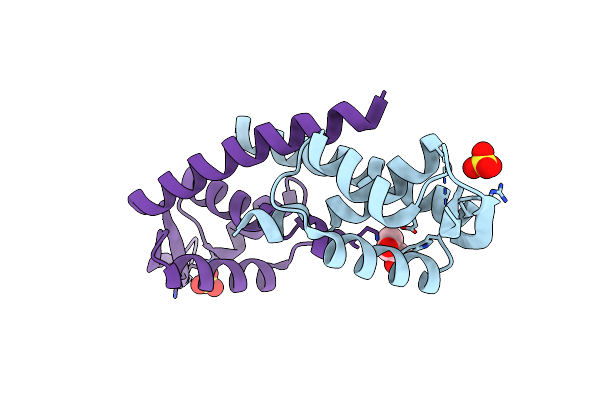 |
Crystal Structure Of Transcriptional Regulator, A Member Of Padr Family, From Enterococcus Faecalis V583
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2009-06-02 Classification: TRANSCRIPTION REGULATOR Ligands: GOL, SO4 |
 |
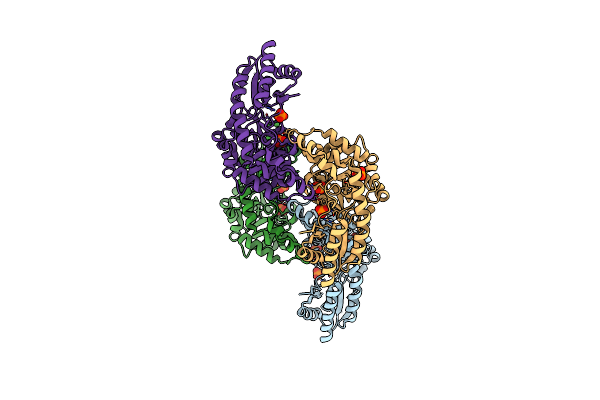
The Crystal Structure Of One Domain Of The Protein With Unknown Function From Methanocaldococcus Jannaschii
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-05-12 Classification: structural genomics, unknown function Ligands: PG6 |
 |
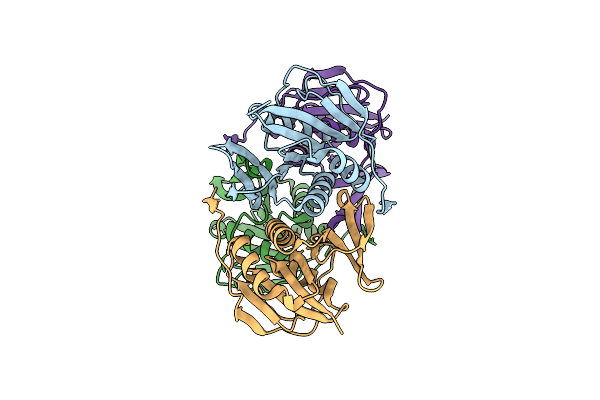
The Crystal Structure Of The Histidinol-Phosphate Aminotransferase From Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-02-17 Classification: TRANSFERASE Ligands: PO4 |
 |
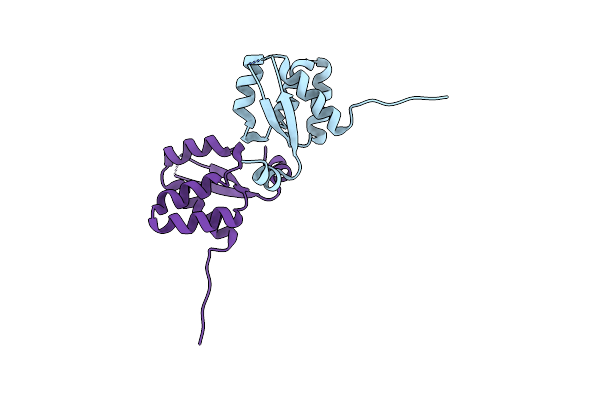
Crystal Structure Of The Electron Transfer Flavoprotein Subunit Alpha Related Protein Ta0212 From Thermoplasma Acidophilum
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2009-01-20 Classification: ELECTRON TRANSPORT |
 |
Structure Of Uncharacterised Protein Rbstp2171 From Bacillus Stearothermophilus
Organism: Bacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2008-12-23 Classification: structural genomics, unknown function |
 |
The Crystal Structure Of A Possible Hxlr Family Transcriptional Factor From Thermoplasma Volcanium Gss1
Organism: Thermoplasma volcanium
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2008-08-05 Classification: TRANSCRIPTION Ligands: ACT |
 |
The Crystal Structure Of A Purr Family Transcriptional Regulator From Vibrio Parahaemolyticus Rimd 2210633
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2008-07-22 Classification: transcription regulator |
 |
Crystal Structure Of Effector Binding Domain Of Central Glycolytic Gene Regulator (Cggr) From Bacillus Subtilis In Complex With Dihydroxyacetone Phosphate
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-07-01 Classification: GENE REGULATION Ligands: 13P |
 |
Crystal Structure Of Effector Binding Domain Of Central Glycolytic Gene Regulator (Cggr) From Bacillus Subtilis In Complex With Effector Fructose-1,6-Bisphosphate
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-07-01 Classification: GENE REGULATION Ligands: FBP, CL, 13P |
 |
Crystal Structure Of Effector Binding Domain Of Central Glycolytic Gene Regulator (Cggr) From Bacillus Subtilis In Complex With Glucose-6-Phosphate
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-07-01 Classification: GENE REGULATION Ligands: BG6 |
 |
Crystal Structure Of Effector Binding Domain Of Central Glycolytic Gene Regulator (Cggr) From Bacillus Subtilis In Complex With Fructose-6-Phosphate
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-07-01 Classification: GENE REGULATION Ligands: F6P, SCN |

