Search Count: 24
 |
Aldehyde Oxidoreductase From Desulfovibrio Gigas (Mop), Soaked With Benzaldehyde
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2014-10-08 Classification: OXIDOREDUCTASE Ligands: FES, PGE, IPA, BCT, MG, CL, PCD, HBX |
 |
Aldehyde Oxidoreductase From Desulfovibrio Gigas (Mop), Soaked With 3- Phenylpropionaldehyde
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-10-08 Classification: OXIDOREDUCTASE Ligands: FES, 3PL, BCT, MG, CL, PCD |
 |
Aldehyde Oxidoreductase From Desulfovibrio Gigas (Mop), Soaked With Trans-Cinnamaldehyde
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2014-10-08 Classification: OXIDOREDUCTASE Ligands: FES, IPA, BCT, MG, CL, PCD, HCI |
 |
Aldehyde Oxidoreductase From Desulfovibrio Gigas (Mop), Soaked With Sodium Dithionite And Sodium Sulfide
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2014-01-15 Classification: OXIDOREDUCTASE Ligands: FES, IPA, BCT, MG, CL, PCD, PEO |
 |
Aldehyde Oxidoreductase From Desulfovibrio Gigas (Mop), Activated With Sodium Dithionite And Sodium Sulfide
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2014-01-15 Classification: OXIDOREDUCTASE Ligands: MG, FES, IPA, BCT, CL, PCD, PEO |
 |
Aldehyde Oxidoreductase From Desulfovibrio Gigas (Mop), Soaked With Hydrogen Peroxide
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-01-15 Classification: OXIDOREDUCTASE Ligands: FES, IPA, BCT, NA, MG, CL, PCD, PEO |
 |
Organism: Desulfovibrio gigas
Method: SOLUTION NMR Release Date: 2012-01-25 Classification: ELECTRON TRANSPORT Ligands: ZN |
 |
Crystal Structure Of The Periplasmic Nitrate Reductase From Cupriavidus Necator
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-04-06 Classification: OXIDOREDUCTASE Ligands: SF4, MOS, MGD, HEC |
 |
Crystal Structure Of Partially Reduced Periplasmic Nitrate Reductase From Cupriavidus Necator Using Ionic Liquids
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2011-04-06 Classification: OXIDOREDUCTASE Ligands: SF4, MOS, MGD, FMT, CL, HEC |
 |
Crystal Structures Of Iron Containing Adenylate Kinase From Desulfovibrio Gigas
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2010-12-15 Classification: TRANSFERASE Ligands: FE, GOL |
 |
Crystal Structures Of Zinc, Cobalt And Iron Containing Adenylate Kinase From Gram-Negative Bacteria Desulfovibrio Gigas
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-12-15 Classification: TRANSFERASE Ligands: CO, TAR |
 |
Crystal Structures Of Zinc Containing Adenylate Kinase From Desulfovibrio Gigas
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-09-22 Classification: TRANSFERASE Ligands: ZN, SRT |
 |
Organism: Desulfovibrio vulgaris
Method: SOLUTION NMR Release Date: 2008-07-08 Classification: ELECTRON TRANSPORT Ligands: ZN |
 |
A New Catalytic Mechanism Of Periplasmic Nitrate Reductase From Desulfovibrio Desulfuricans Atcc 27774 From Crystallographic And Epr Data And Based On Detailed Analysis Of The Sixth Ligand
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2008-03-18 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, UNX |
 |
A New Catalytic Mechanism Of Periplasmic Nitrate Reductase From Desulfovibrio Desulfuricans Atcc 27774 From Crystallographic And Epr Data And Based On Detailed Analysis Of The Sixth Ligand
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-03-18 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, UNX |
 |
A New Catalytic Mechanism Of Periplasmic Nitrate Reductase From Desulfovibrio Desulfuricans Atcc 27774 From Crystallographic And Epr Data And Based On Detailed Analysis Of The Sixth Ligand
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-03-18 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, UNX |
 |
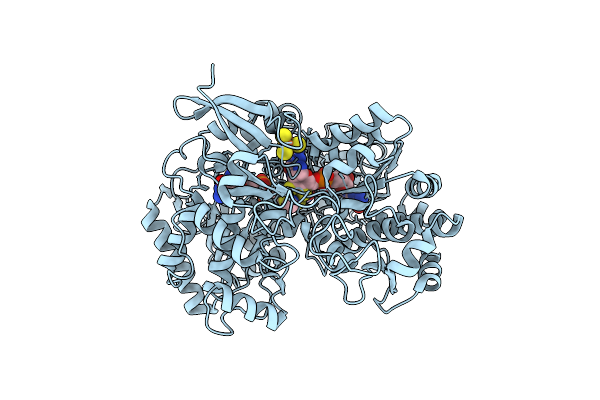
A New Catalytic Mechanism Of Periplasmic Nitrate Reductase From Desulfovibrio Desulfuricans Atcc 27774 From Crystallographic And Epr Data And Based On Detailed Analysis Of The Sixth Ligand
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2008-03-18 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, UNX, NO2, NO3 |
 |
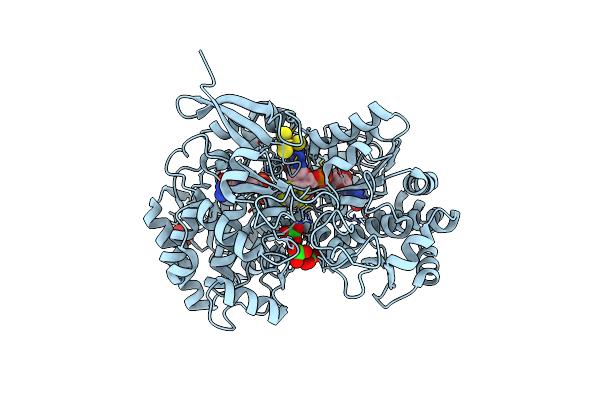
A New Catalytic Mechanism Of Periplasmic Nitrate Reductase From Desulfovibrio Desulfuricans Atcc 27774 From Crystallographic And Epr Data And Based On Detailed Analysis Of The Sixth Ligand
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2008-03-18 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, CYN, NO2 |
 |
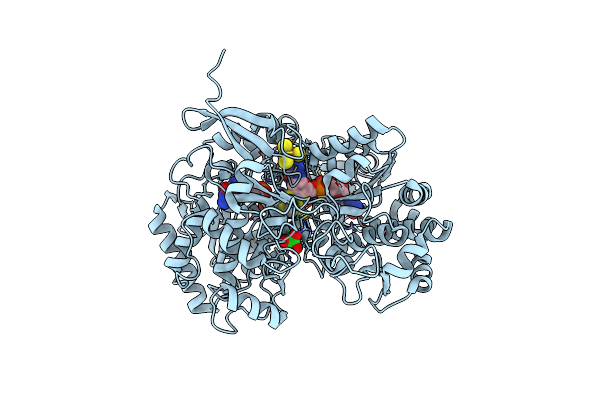
A New Catalytic Mechanism Of Periplasmic Nitrate Reductase From Desulfovibrio Desulfuricans Atcc 27774 From Crystallographic And Epr Data And Based On Detailed Analysis Of The Sixth Ligand
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2008-03-18 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, UNX, LCP |
 |
A New Catalytic Mechanism Of Periplasmic Nitrate Reductase From Desulfovibrio Desulfuricans Atcc 27774 From Crystallographic And Epr Data And Based On Detailed Analysis Of The Sixth Ligand
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-03-18 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, UNX, LCP |

