Search Count: 14
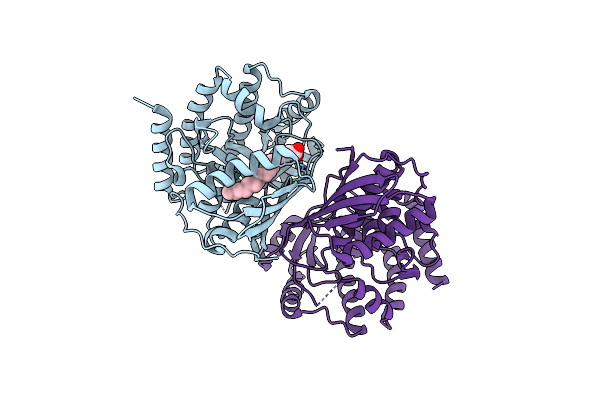 |
Crystal Structure Of Mevalonate 3,5-Bisphosphate Decarboxylase From Picrophilus Torridus
Organism: Picrophilus torridus dsm 9790
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2021-12-22 Classification: LYASE Ligands: OLA |
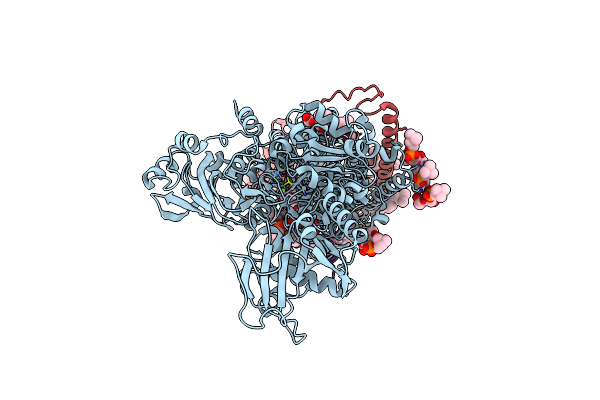 |
Crystal Structure Of The Na+,K+-Atpase In The E2P State With Bound Mg2+ (P4(3)2(1)2 Symmetry)
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2021-01-27 Classification: TRANSPORT PROTEIN Ligands: MG, CLR, PCW, NAG |
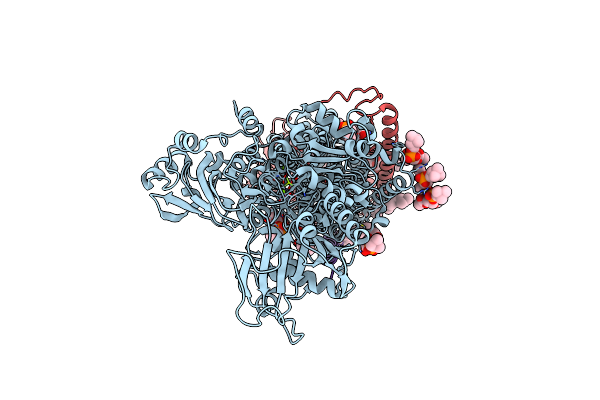 |
Crystal Structure Of The Na+,K+-Atpase In The E2P State With Bound Mg2+ And Anthroylouabain (P4(3)2(1)2 Symmetry)
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2021-01-27 Classification: TRANSPORT PROTEIN Ligands: MG, CLR, PCW, H0C, NAG |
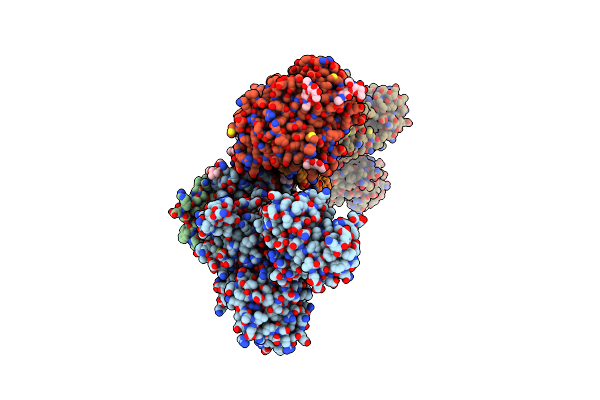 |
Crystal Structure Of The Na+,K+-Atpase In The E2P State With Bound Mg2+ And Anthroylouabain (P2(1)2(1)2(1) Symmetry)
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:3.65 Å Release Date: 2021-01-27 Classification: TRANSPORT PROTEIN Ligands: MG, NA, CLR, PCW, H0C, NAG |
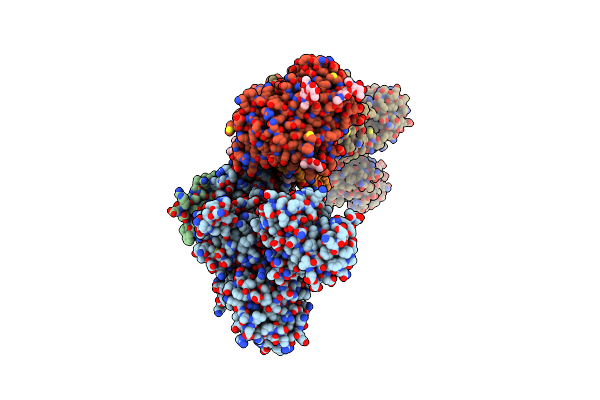 |
Crystal Structure Of The Na+,K+-Atpase In The E2P State With Bound One Mg2+ And One Rb+ In The Presence Of Bufalin
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2021-01-27 Classification: TRANSPORT PROTEIN Ligands: MG, NA, RB, CLR, PCW, BUF, NAG |
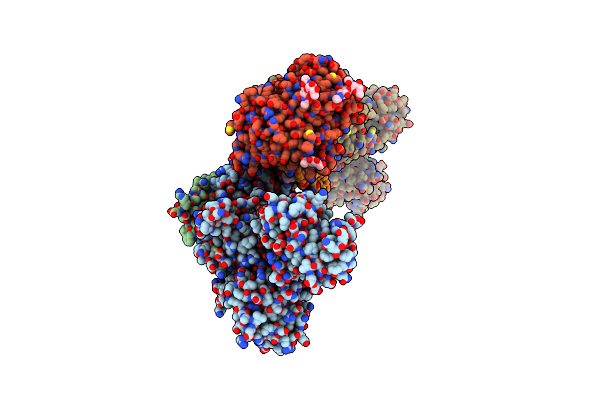 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:4.62 Å Release Date: 2021-01-27 Classification: MEMBRANE PROTEIN Ligands: MG, NA, CLR, PCW, NAG |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:3.46 Å Release Date: 2021-01-27 Classification: MEMBRANE PROTEIN Ligands: MG, NA, CLR, PCW, DGX, NAG |
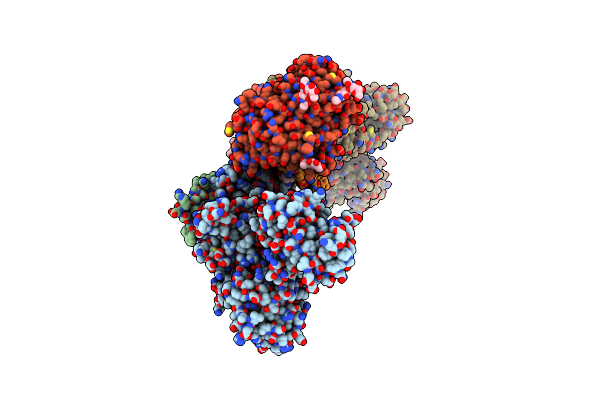 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:3.72 Å Release Date: 2021-01-27 Classification: MEMBRANE PROTEIN Ligands: MG, NA, PCW, F9R, CLR, NAG |
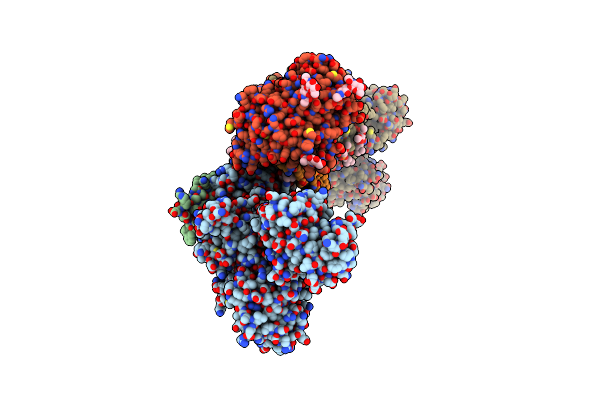 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2021-01-27 Classification: MEMBRANE PROTEIN Ligands: MG, NA, PCW, E4R, CLR, NAG |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2021-01-27 Classification: MEMBRANE PROTEIN Ligands: MG, NA, PCW, BUF, CLR, NAG |
 |
Crystal Structure Of Sulfolobus Solfataricus Diphosphomevalonate Decarboxylase In Complex With Atp-Gamma-S
Organism: Sulfolobus solfataricus (strain atcc 35092 / dsm 1617 / jcm 11322 / p2)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-12-28 Classification: LYASE Ligands: DP6, AGS, AMP, SO4 |
 |
Crystal Structure Of Sulfolobus Solfataricus Diphosphomevalonate Decarboxylase In Complex With Adp
Organism: Sulfolobus solfataricus (strain atcc 35092 / dsm 1617 / jcm 11322 / p2)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-12-28 Classification: LYASE Ligands: DP6, ADP, AMP, SO4 |
 |
Diphosphomevalonate Decarboxylase From The Sulfolobus Solfataricus, Space Group H32
Organism: Sulfolobus solfataricus (strain atcc 35092 / dsm 1617 / jcm 11322 / p2)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-09-16 Classification: LYASE Ligands: PO4, NA, GOL |
 |
Diphosphomevalonate Decarboxylase From The Sulfolobus Solfataricus, Space Group P21
Organism: Sulfolobus solfataricus (strain atcc 35092 / dsm 1617 / jcm 11322 / p2)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-09-16 Classification: LYASE Ligands: SO4 |

