Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: DE NOVO PROTEIN Ligands: PGE, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: DE NOVO PROTEIN Ligands: GOL, SO4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: DE NOVO PROTEIN Ligands: ACT, GOL, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: DE NOVO PROTEIN Ligands: 2PE |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: DE NOVO PROTEIN Ligands: MG, AE4 |
 |
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-01-01 Classification: LIGASE Ligands: 1PE, NA, GOL, EDO |
 |
Solution Nmr Structure Of The Myristoylated Feline Immunodeficiency Virus Matrix Protein
Organism: Feline immunodeficiency virus
Method: SOLUTION NMR Release Date: 2020-07-22 Classification: VIRAL PROTEIN Ligands: MYR |
 |
Solution Nmr Structure Of The Unmyristoylated Feline Immunodeficiency Virus Matrix Protein
Organism: Feline immunodeficiency virus
Method: SOLUTION NMR Release Date: 2020-07-22 Classification: VIRAL PROTEIN |
 |
Solution Nmr Structure Of The G4L/Q5K/G6S (Nos) Unmyristoylated Feline Immunodeficiency Virus Matrix Protein
Organism: Feline immunodeficiency virus
Method: SOLUTION NMR Release Date: 2020-07-22 Classification: VIRAL PROTEIN |
 |
Organism: Mus musculus, Homo sapiens
Method: SOLUTION NMR Release Date: 2017-12-06 Classification: CELL ADHESION |
 |
Structure Of Porcine Trypsin Complexed With Bdellastasin, An Antistasin-Type Inhibitor
Organism: Hirudo medicinalis, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2001-03-02 Classification: HYDROLASE/INHIBITOR |
 |
Organism: Hirudo medicinalis, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2000-08-03 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA |
 |
Organism: Hirudo medicinalis, Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2000-08-03 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
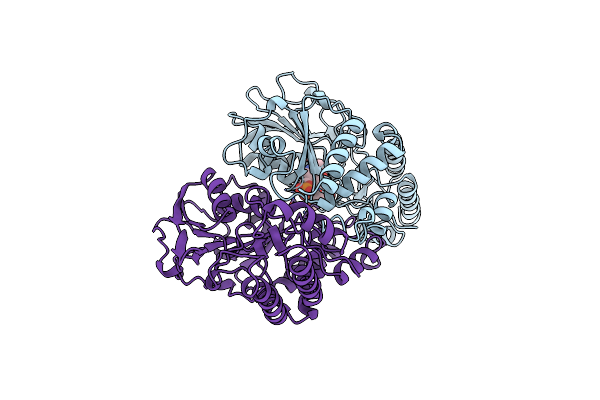
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1998-11-04 Classification: AMINOTRANSFERASE |
 |
The Structure Of Phosphoserine Aminotransferase From E. Coli In Complex With Alpha-Methyl-L-Glutamate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 1998-11-04 Classification: AMINOTRANSFERASE Ligands: PLP, GAM |
 |
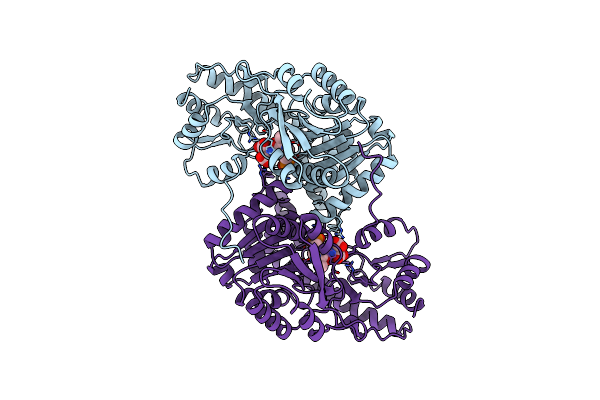
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1998-09-09 Classification: TRANSFERASE Ligands: PLP |
 |
Crystal Structures Of Escherichia Coli Aspartate Aminotransferase In Two Conformations: Comparison Of An Unliganded Open And Two Liganded Closed Forms
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 1994-01-31 Classification: AMINOTRANSFERASE Ligands: PLA |
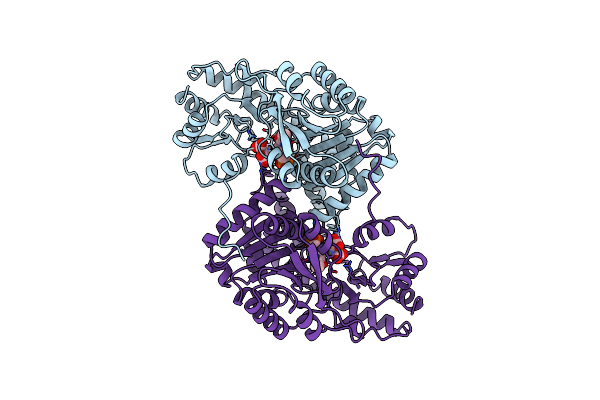 |
Crystal Structures Of Escherichia Coli Aspartate Aminotransferase In Two Conformations: Comparison Of An Unliganded Open And Two Liganded Closed Forms
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 1994-01-31 Classification: AMINOTRANSFERASE Ligands: PLP, MAE |
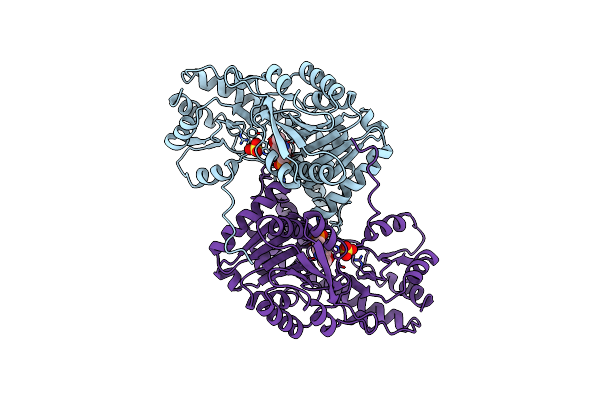 |
Crystal Structures Of Escherichia Coli Aspartate Aminotransferase In Two Conformations: Comparison Of An Unliganded Open And Two Liganded Closed Forms
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1994-01-31 Classification: AMINOTRANSFERASE Ligands: SO4, PLP |