Search Count: 15
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: TRANSPORT PROTEIN Ligands: FUN |
 |
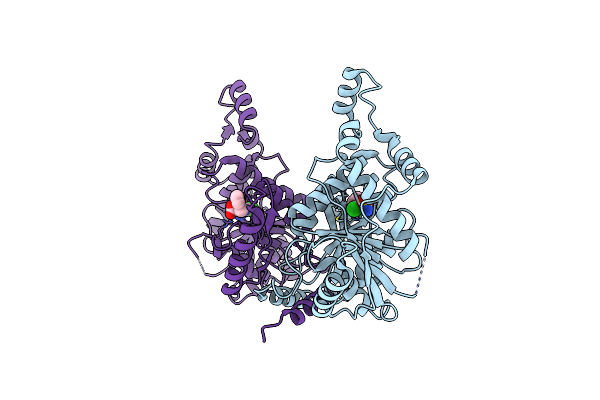
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: MEMBRANE PROTEIN Ligands: CL, K |
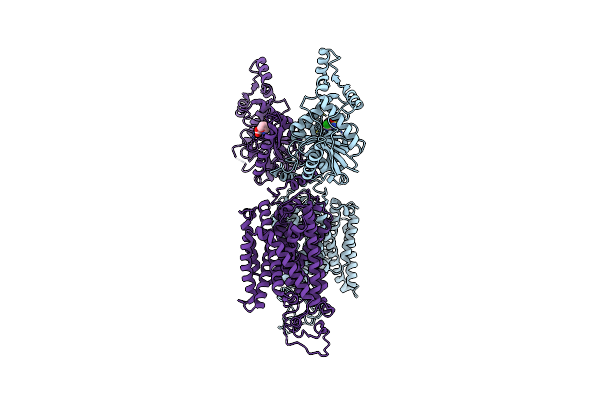 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: MEMBRANE PROTEIN Ligands: FUN |
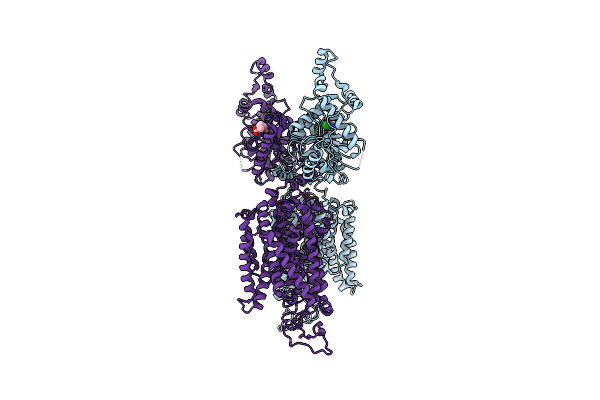 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: MEMBRANE PROTEIN Ligands: FUN |
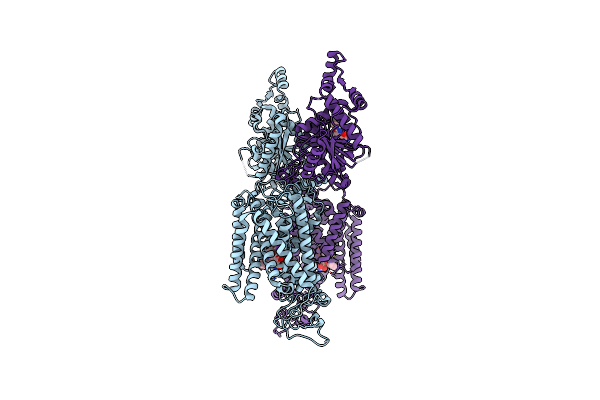 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: MEMBRANE PROTEIN, Transport Protein Ligands: 82U, K |
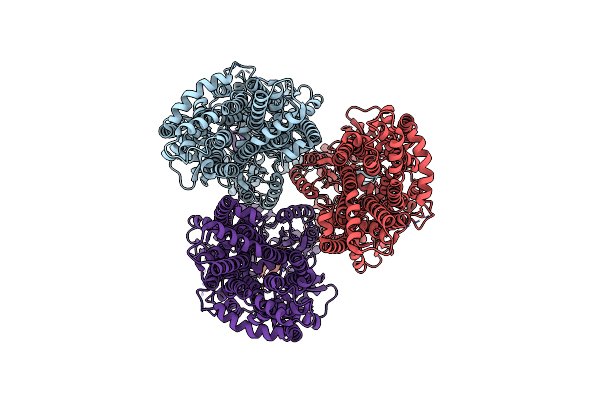 |
Cryo-Electron Microscopy Structure Of The Heavy Metal Efflux Pump Cusa In The Symmetric Closed State
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN |
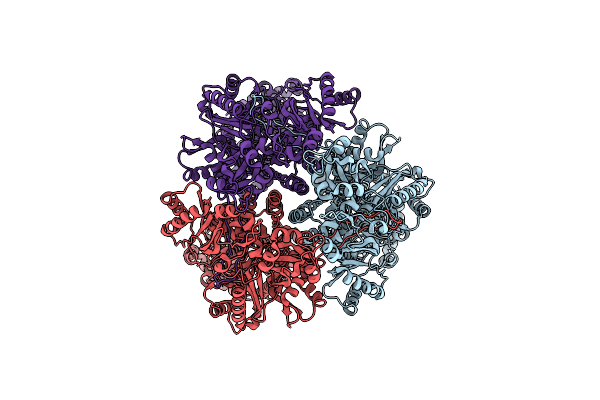 |
Cryo-Electron Microscopy Structure Of The Heavy Metal Efflux Pump Cusa In A Homogeneous Binding Copper(1) State
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN Ligands: CU1 |
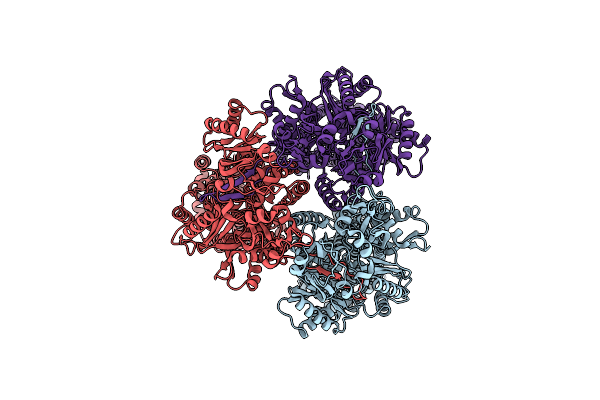 |
Cryo-Electron Microscopy Structure Of The Heavy Metal Efflux Pump Cusa In A Heterogeneous 1 Open And 2 Closed Protomer Conformation
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN/ TRANSPORT PROTEIN Ligands: CU1 |
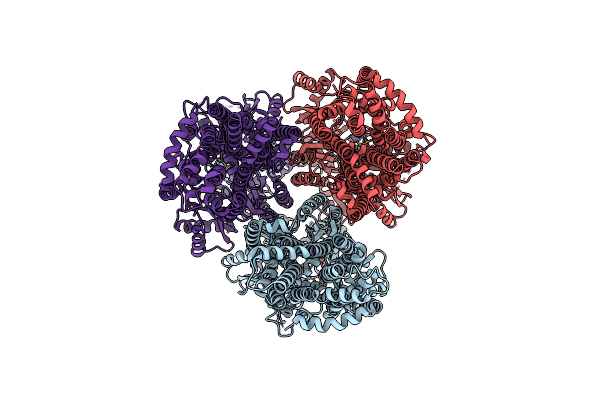 |
Cryo-Electron Microscopy Structure Of The Heavy Metal Efflux Pump Cusa In A Heterogeneous 2 Open And 1 Closed Protomer Conformation
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN Ligands: CU1 |
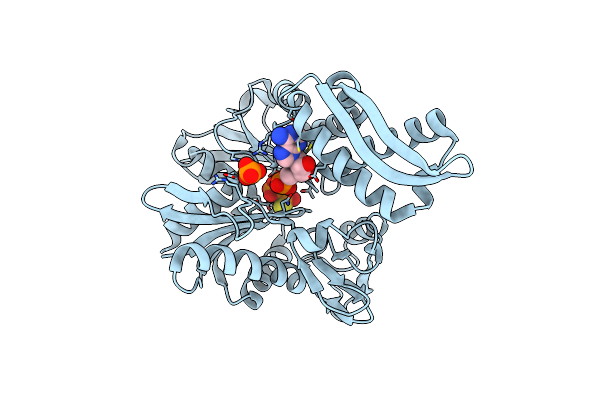 |
Structure Of Mortalin-Nbd With Adenosine-5'-Monophosphate And Thiodiphosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-07-08 Classification: CHAPERONE Ligands: AMP, PIS, PO4 |
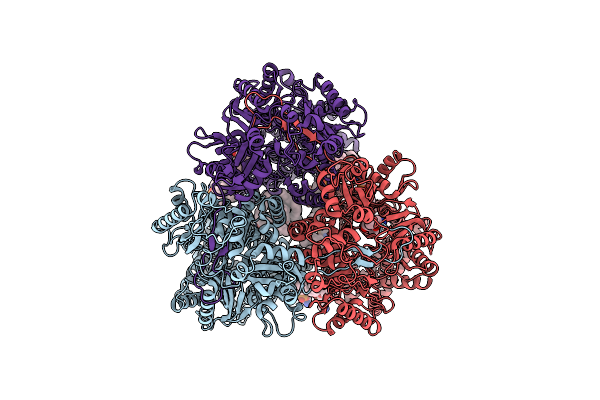 |
Cryo-Electron Microscopy Structures Of A Gonococcal Multidrug Efflux Pump Illuminate A Mechanism Of Drug Recognition With Ampicillin
Organism: Neisseria gonorrhoeae
Method: ELECTRON MICROSCOPY Release Date: 2020-07-01 Classification: MEMBRANE PROTEIN Ligands: PTY, AIX |
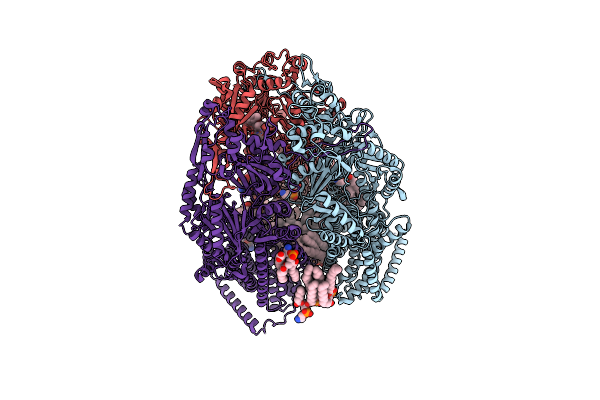 |
Cryo-Electron Microscopy Structures Of A Gonococcal Multidrug Efflux Pump Illuminate A Mechanism Of Erythromycin Drug Recognition
Organism: Neisseria gonorrhoeae
Method: ELECTRON MICROSCOPY Release Date: 2020-07-01 Classification: MEMBRANE PROTEIN Ligands: PTY, ERY |
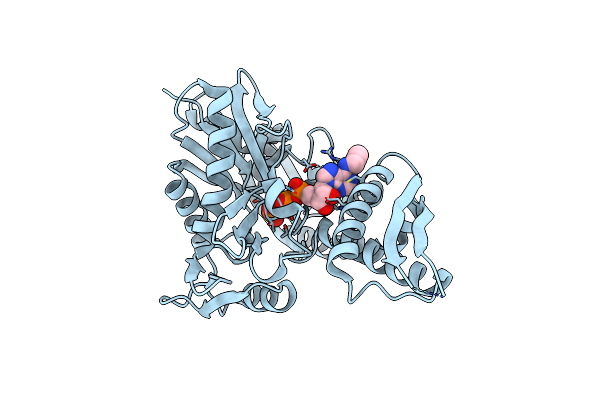 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-06-19 Classification: CHAPERONE Ligands: NO7, PO4, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2019-04-17 Classification: CHAPERONE Ligands: ADP, PO4, MG, GOL |
 |
Crystal Structure Of The Human Mortalin (Grp75) Atpase Domain In The Apo Form
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-04-02 Classification: SIGNALING PROTEIN |

