Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Structure Of The Human Systemic Rnai Defective Transmembrane Protein 1 (Hsidt1)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Aequorea victoria, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: TRANSFERASE Ligands: ACO, NAG |
 |
Organism: Rotavirus a
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: VIRAL PROTEIN |
 |
Organism: Rotavirus a
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: VIRAL PROTEIN Ligands: CA, ZN |
 |
Organism: Rotavirus a
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: VIRAL PROTEIN |
 |
Organism: Methylococcus capsulatus
Method: ELECTRON MICROSCOPY Release Date: 2023-01-25 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Organism: Methylococcus capsulatus
Method: ELECTRON MICROSCOPY Release Date: 2023-01-25 Classification: OXIDOREDUCTASE Ligands: FE |
 |
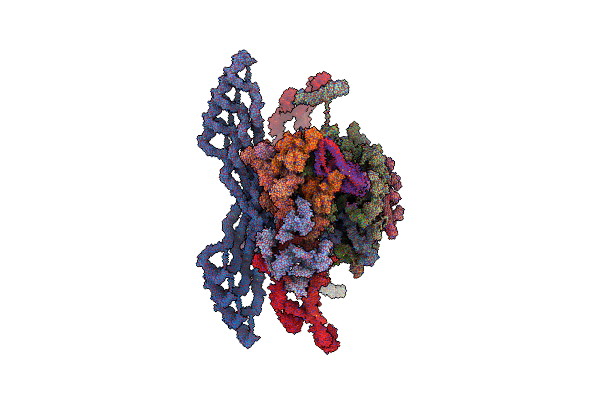
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-09-21 Classification: TRANSPORT PROTEIN |
 |
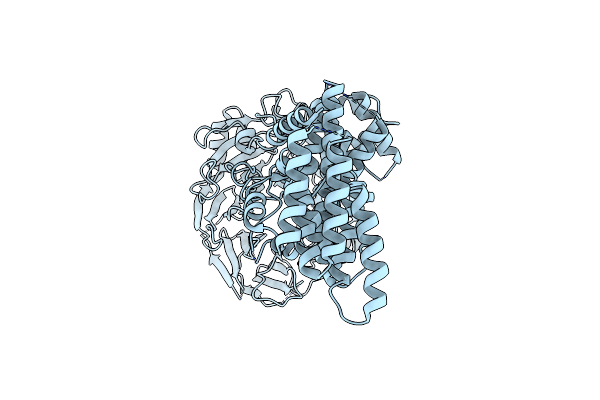
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-06-08 Classification: STRUCTURAL PROTEIN |
 |
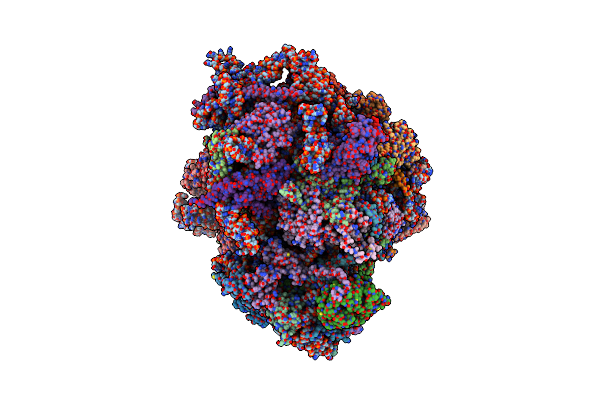
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2020-08-26 Classification: RIBOSOME Ligands: ZN |
 |
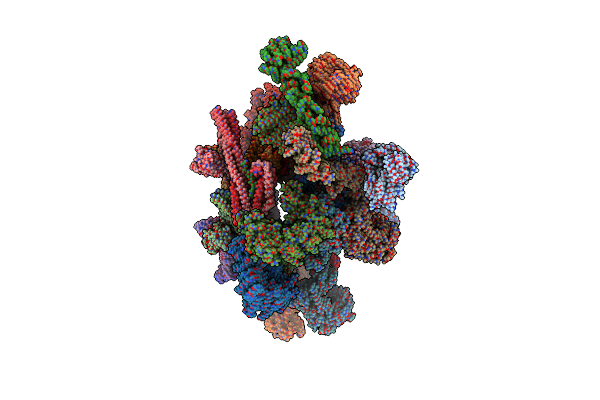
Cryo-Em Structure Of K63 Ubiquitinated Yeast Translocating Ribosome Under Oxidative Stress
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2020-08-26 Classification: RIBOSOME Ligands: ZN |
 |
Composite Structure Of The Inner Ring Of The Human Nuclear Pore Complex (32 Copies Of Nup205)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2016-04-27 Classification: TRANSPORT PROTEIN |
 |
Alternative Composite Structure Of The Inner Ring Of The Human Nuclear Pore Complex (16 Copies Of Nup188, 16 Copies Of Nup205)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2016-04-27 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:23.00 Å Release Date: 2015-09-30 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-03-19 Classification: CELL CYCLE Ligands: CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-03-19 Classification: CELL CYCLE Ligands: CL, GOL |