Search Count: 96
 |
Gh191 Family Alpha-Galactosaminidase From Environmental Sample (99.2% Identity To Myxococcus Fulvus Enzyme)
Organism: Environmental samples
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Fusarium solani
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: EDO, PEG |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: HYDROLASE |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: HYDROLASE |
 |
Organism: Terribacillus saccharophilus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: ACT |
 |
Organism: Terribacillus saccharophilus
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: GC2, MPD |
 |
Organism: Lactiplantibacillus paraplantarum
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: GC2, EDO, PEG |
 |
Dispersin From Terribacillus Saccharophilus Dispts2 In Complex With Nag-Thiazoline
Organism: Terribacillus saccharophilus
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: NGT |
 |
Crystal Structure Of Susa Amylase From Bacteroides Thetaiotaomicron Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, OC9, IMD, CA |
 |
Crystal Structure Of Susg From Bacteroides Thetaiotaomicron Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, OC9, A1ILG, CA, IMD, ACT |
 |
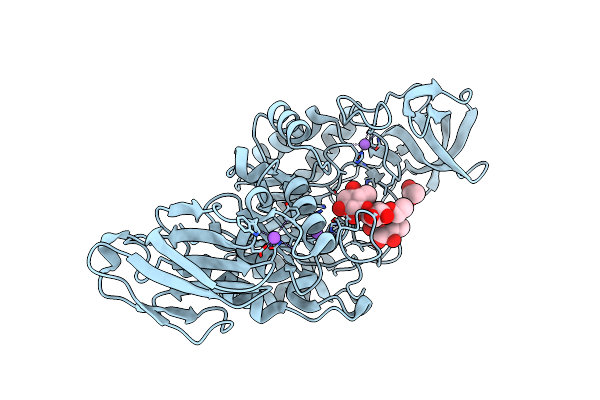
Crystal Structure Of Amylase 5 (Amy5) From Ruminococcus Bromii Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Ruminococcus bromii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, A1IHI, CA |
 |
Crystal Structure Of K38 Amylase From Bacillus Sp. Strain Ksm-K38 Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Bacillus sp. ksm-k38
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, OC9, ACT, NA |
 |
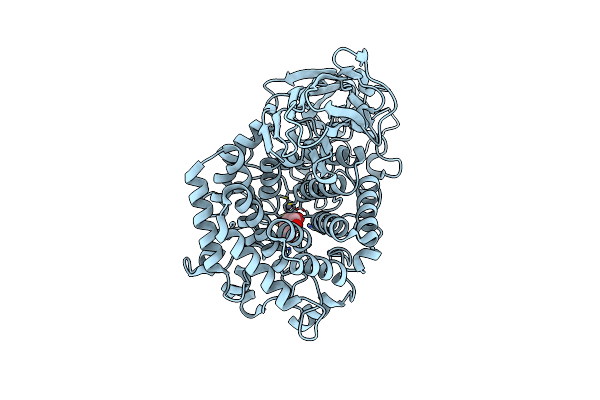
Crystal Structure Of Mgat5 Bump-And-Hole Mutant In Complex With Udp And M592
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-10-02 Classification: TRANSFERASE Ligands: UDP, NAG, MAN, SO4 |
 |
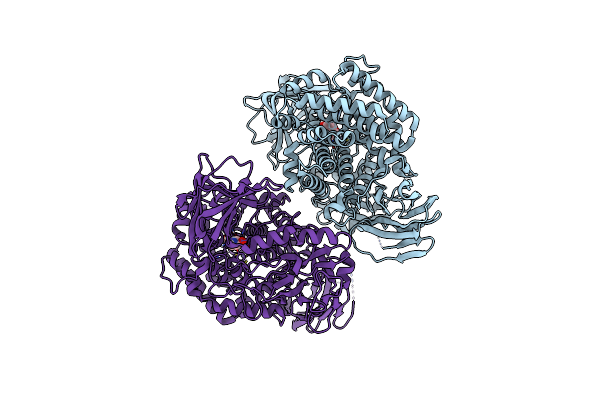
Beta-L-Arabinofurano-Cyclitol Aziridines Are Cysteine-Directed Broad-Spectrum Inhibitors And Activity-Based Probes For Retaining Beta-L-Arabinofuranosidases
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-12-13 Classification: HYDROLASE Ligands: ZN, UI5 |
 |
Gh146 Beta-L-Arabinofuranosidase From Bacteroides Thetaioatomicron In Complex With Beta-L-Arabinofurano Cyclophellitol Aziridine
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-12-13 Classification: HYDROLASE Ligands: UI5, ZN, UHU |
 |
Organism: Kionochaeta sp.
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-07-19 Classification: HYDROLASE Ligands: CD |
 |
Sh3-Like Cell Wall Binding Domain Of The Gh24 Family Muramidase From Trichophaea Saccata In Complex With Triglycine
Organism: Trichophaea saccata, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2023-07-19 Classification: HYDROLASE Ligands: EDO, ZN |
 |
Organism: Penicillium virgatum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-07-19 Classification: HYDROLASE Ligands: EDO, ZN |
 |
Organism: Thermothielavioides terrestris
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2023-07-19 Classification: HYDROLASE Ligands: EDO, ZN |
 |
Gh24 Family Muramidase From Trichophaea Saccata With An Sh3-Like Cell Wall Binding Domain
Organism: Trichophaea saccata
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2023-07-19 Classification: HYDROLASE Ligands: K |

