Search Count: 26
 |
Organism: Mycolicibacterium chubuense, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: 6OU |
 |
Organism: Mycolicibacterium chubuense
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: A1A7X |
 |
Organism: Mycobacteroides abscessus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: A1A8B, DSL |
 |
Organism: Mycobacteroides abscessus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN |
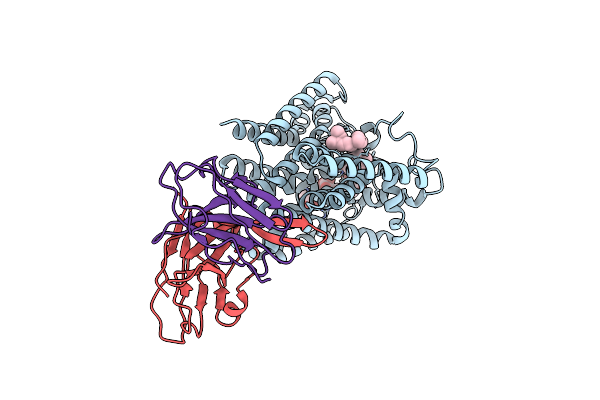 |
Organism: Mycobacteroides abscessus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: A1A8B, DSL |
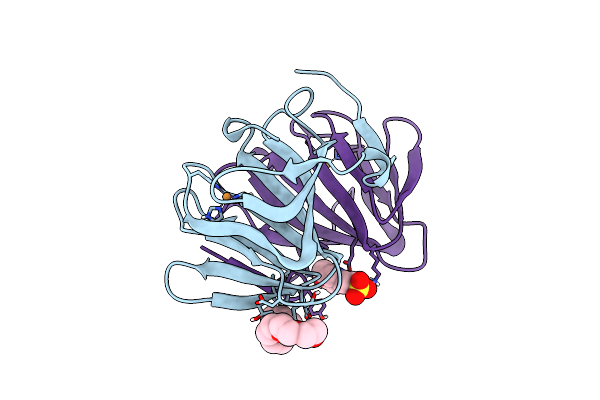 |
Crystal Structure Of A Cupin Protein (Tm1459, I49C-4Py/H52A/C106D Mutant) In Copper (Cu) Substituted Form
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2024-09-04 Classification: METAL BINDING PROTEIN Ligands: P6G, CU, MES |
 |
Crystal Structure Of A Cupin Protein (Tm1459, I49C-4Py/H52A/H54A/C106D Mutant) In Copper (Cu) Substituted Form
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2024-09-04 Classification: METAL BINDING PROTEIN Ligands: P6G, CU, KN6 |
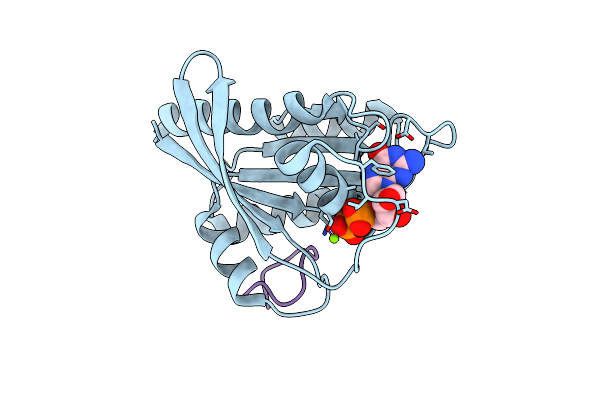 |
Human K-Ras G12D (Gdp-Bound) In Complex With Cyclic Peptide Inhibitor Ap8784
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-07-26 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: GDP, MG, IOD |
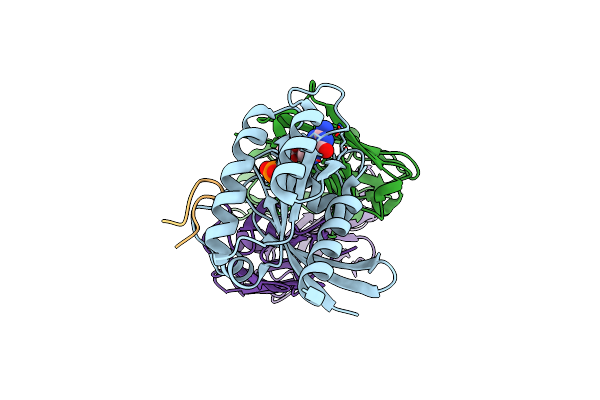 |
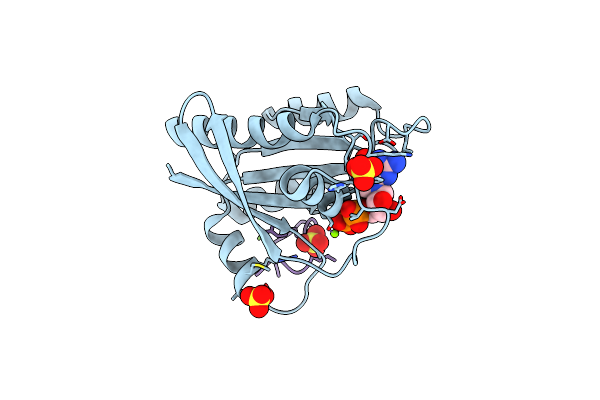
Human K-Ras G12D (Gdp-Bound) In Complex With Cyclic Peptide Inhibitor Luna18 And Ka30L Fab
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-07-26 Classification: SIGNALING PROTEIN Ligands: GDP, MG |
 |
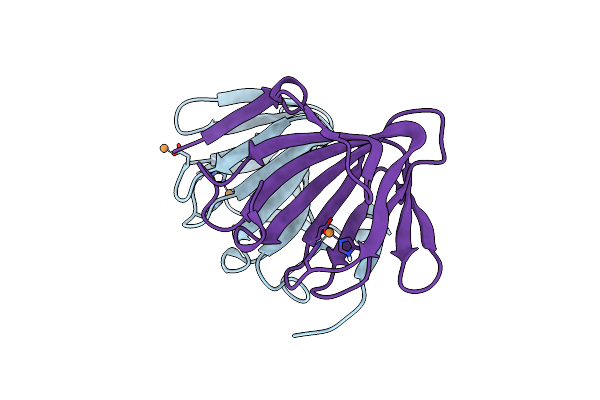
Human K-Ras G12D (Gdp-Bound) In Complex With Cyclic Peptide Inhibitor Ap10343
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2023-07-26 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: GDP, MG, SO4 |
 |
Crystal Structure Of A Cupin Protein (Tm1459, H52A/H58E Mutant) In Copper (Cu) Substituted Form
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2023-06-14 Classification: METAL BINDING PROTEIN Ligands: CU |
 |
Crystal Structure Of A Cupin Protein (Tm1459, H52A/H58E/F104W Mutant) In Copper (Cu) Substituted Form
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2023-06-14 Classification: METAL BINDING PROTEIN Ligands: CU |
 |
Crystal Structure Of A Cupin Protein (Tm1459, H52A/H58Q Mutant) In Copper (Cu) Substituted Form
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2023-06-14 Classification: METAL BINDING PROTEIN Ligands: CU |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2020-01-01 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2018-12-12 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2018-12-12 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO |
 |
Organism: Felis catus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2018-04-04 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of Cyano-Cobalt(Iii) Tetradehydrocorrin In The Heme Pocket Of Horse Heart Myoglobin
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-01-20 Classification: OXYGEN TRANSPORT Ligands: J1R, J1S, CYN, SO4 |
 |
Crystal Structure Of Aqua-Cobalt(Iii) Tetradehydrocorrin In The Heme Pocket Of Horse Heart Myoglobin
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2016-01-20 Classification: OXYGEN TRANSPORT Ligands: J1R, J1S, SO4, GOL |
 |
Crystal Structure Of Horse Heart Myoglobin Reconstituted With Cobalt(Ii) Tetradehydrocorrin
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2014-12-03 Classification: OXYGEN TRANSPORT Ligands: J1R, J1S, SO4, GOL |

