Search Count: 28
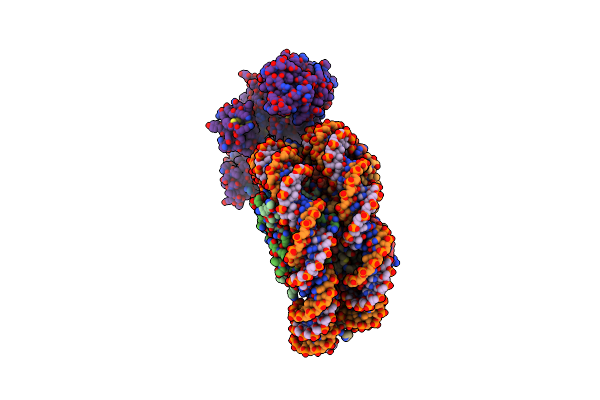 |
Cryo-Em Structure Of The P300 Catalytic Core Bound To The H4K12Ack16Ac Nucleosome, Class 1 (3.2 Angstrom Resolution)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: TRANSFERASE/DNA |
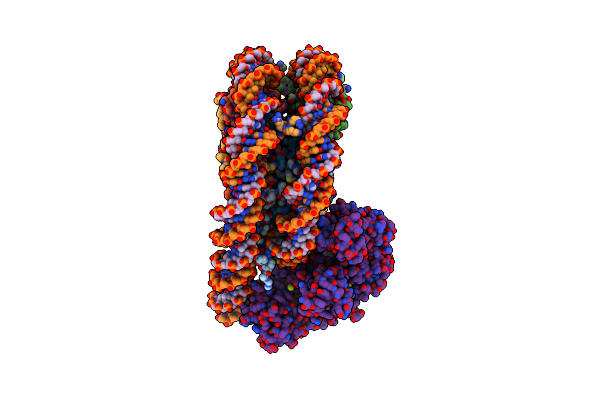 |
Cryo-Em Structure Of The P300 Catalytic Core Bound To The H4K12Ack16Ac Nucleosome, Class 2 (3.9 Angstrom Resolution)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: TRANSFERASE/DNA |
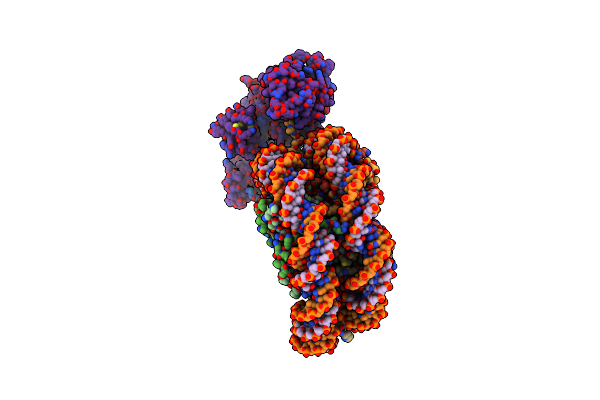 |
Cryo-Em Structure Of The P300 Catalytic Core Bound To The H4K12Ack16Ac Nucleosome, Class 1 (4.7 Angstrom Resolution)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: TRANSFERASE/DNA |
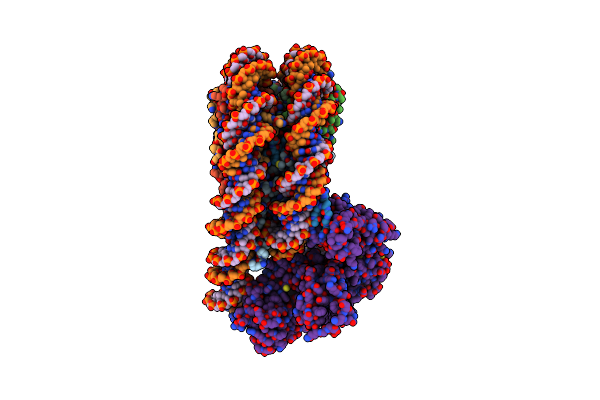 |
Cryo-Em Structure Of The P300 Catalytic Core Bound To The H4K12Ack16Ac Nucleosome, Class 2 (4.8 Angstrom Resolution)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: TRANSFERASE/DNA |
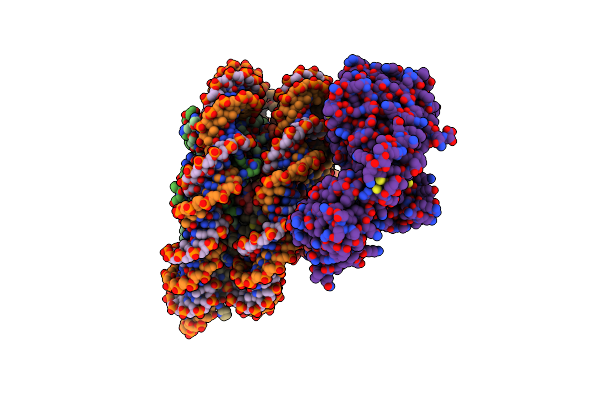 |
Cryo-Em Structure Of The P300 Catalytic Core Bound To The H4K12Ack16Ac Nucleosome, Class 4 (4.5 Angstrom Resolution)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: TRANSFERASE/DNA |
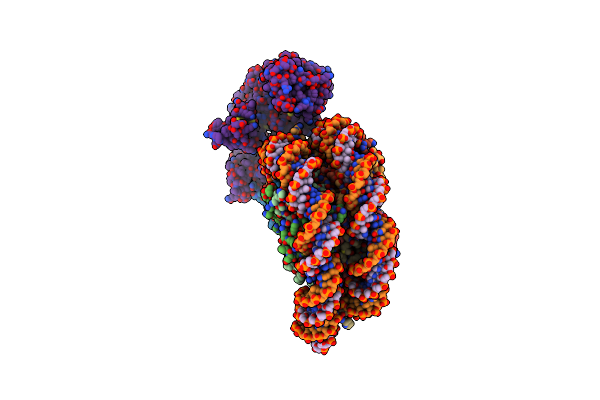 |
Cryo-Em Structure Of The Cbp Catalytic Core Bound To The H4K12Ack16Ac Nucleosome, Class 1
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: TRANSFERASE/DNA |
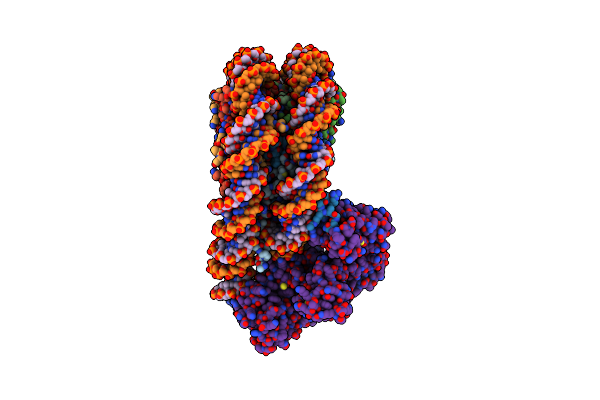 |
Cryo-Em Structure Of The Cbp Catalytic Core Bound To The H4K12Ack16Ac Nucleosome, Class 2
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: TRANSFERASE/DNA |
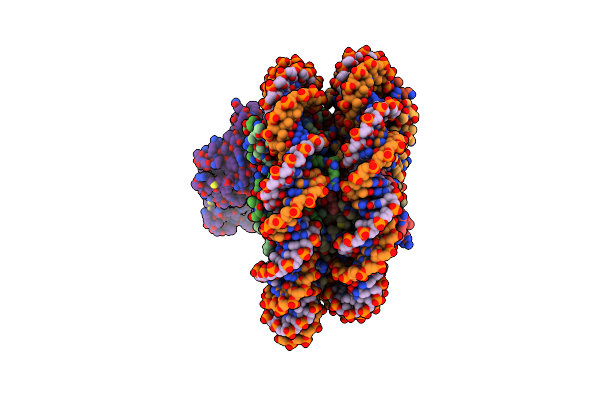 |
Cryo-Em Structure Of The Cbp Catalytic Core Bound To The H4K12Ack16Ac Nucleosome, Class 3
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: TRANSFERASE/DNA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2022-09-07 Classification: PLANT PROTEIN Ligands: TLA, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-04-06 Classification: NUCLEAR PROTEIN Ligands: GOL, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-04-06 Classification: NUCLEAR PROTEIN Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2022-04-06 Classification: NUCLEAR PROTEIN Ligands: GOL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2021-01-27 Classification: HYDROLASE |
 |
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2011-12-07 Classification: TRANSFERASE Ligands: ZN, SO4 |
 |
Crystal Structure Of A Rok Family Glucokinase From Streptomyces Griseus In Complex With Glucose And Amppnp
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2011-12-07 Classification: TRANSFERASE Ligands: ZN, NA, BGC, ANP |
 |
Crystal Structure Of A Rok Family Glucokinase From Streptomyces Griseus In Complex With Glucose
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2011-12-07 Classification: TRANSFERASE Ligands: BGC, ZN, K |
 |
Crystal Structure Of The C-Terminal Gaf Domain Of Human Phosphodiesterase 10A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-04-29 Classification: HYDROLASE Ligands: CMP |
 |
Crystal Structure Of Rrm-Domain Derived From Human Putative Rna-Binding Protein 11
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2008-04-22 Classification: RNA BINDING PROTEIN |
 |
Crystal Structure Of Glutathione Transferase Zeta 1-1 (Maleylacetoacetate Isomerase) From Mus Musculus (Form-1 Crystal)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2006-01-10 Classification: Isomerase, TRANSFERASE Ligands: GSH, GOL |
 |
Crystal Structure Of Glutathione Transferase Zeta 1-1 (Maleylacetoacetate Isomerase) From Mus Musculus (Form-2 Crystal)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-01-10 Classification: Isomerase, TRANSFERASE |

