Search Count: 368
 |
Organism: Kitasatospora
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: LIGASE Ligands: A1L46, GOL |
 |
Organism: Kitasatospora
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: LIGASE Ligands: TRP |
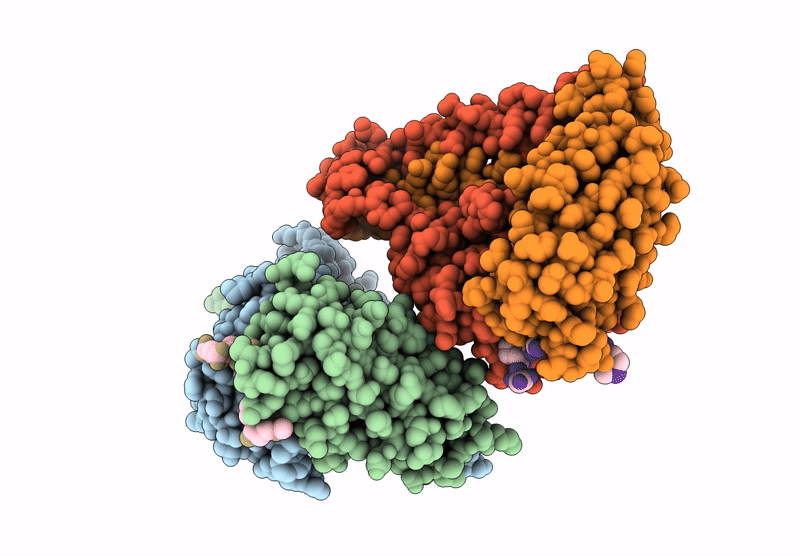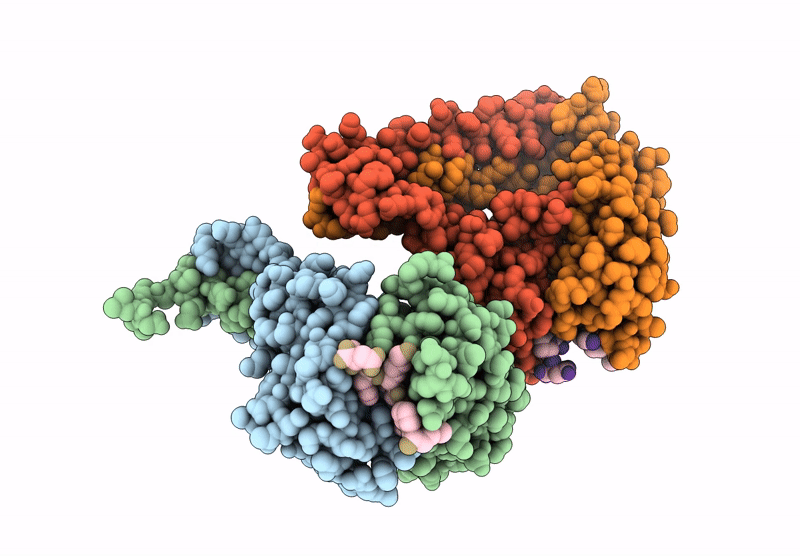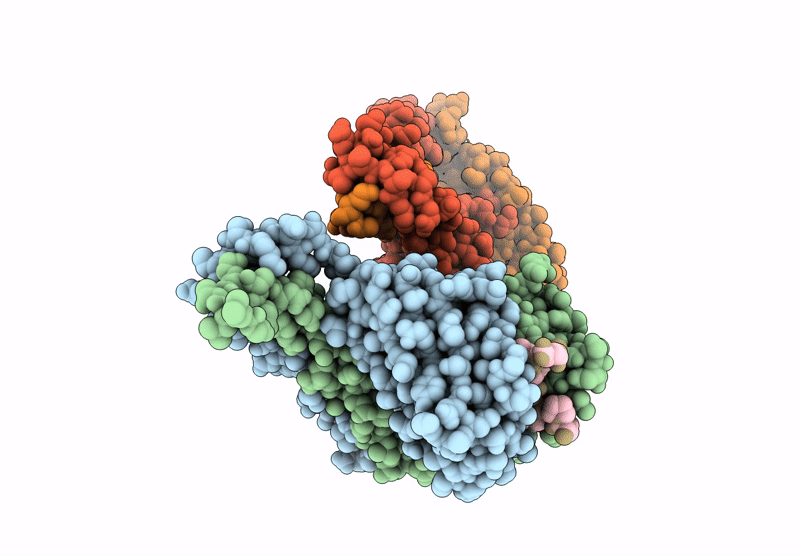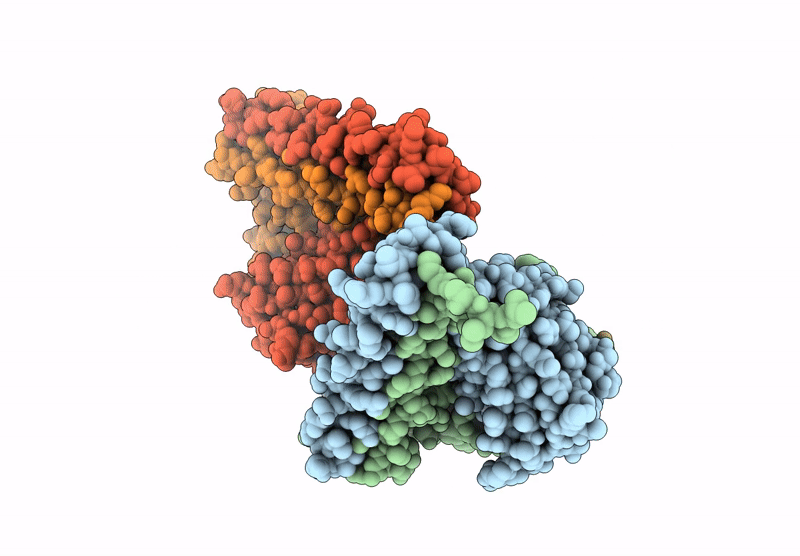 |
Crystal Structure Of Cancer-Specific Anti-Her2 Antibody H2Mab-250 In Complex With Epitope Peptide
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: IMMUNE SYSTEM |
 |
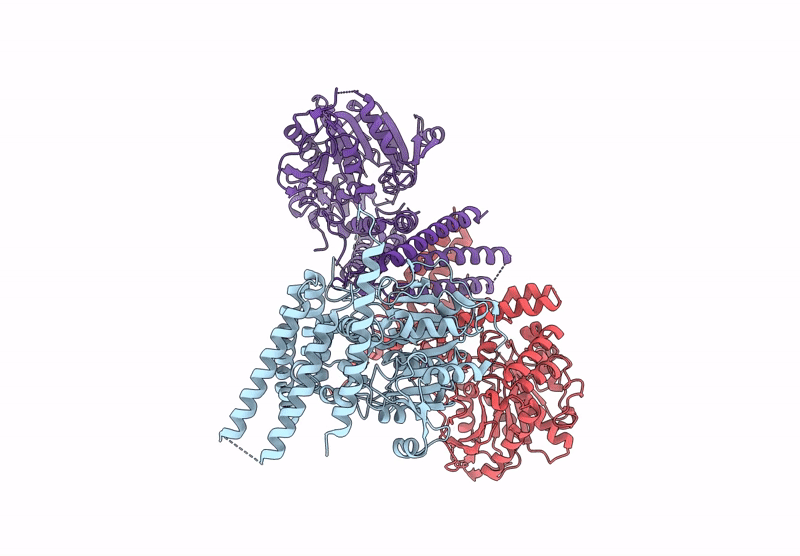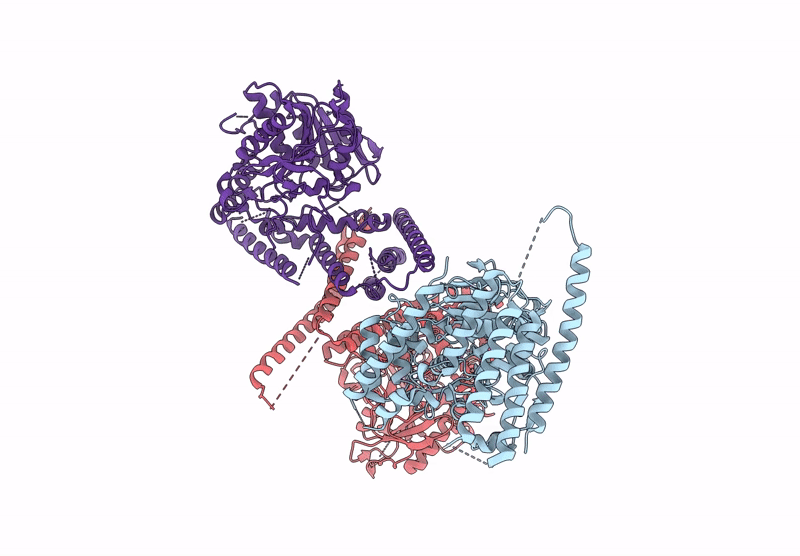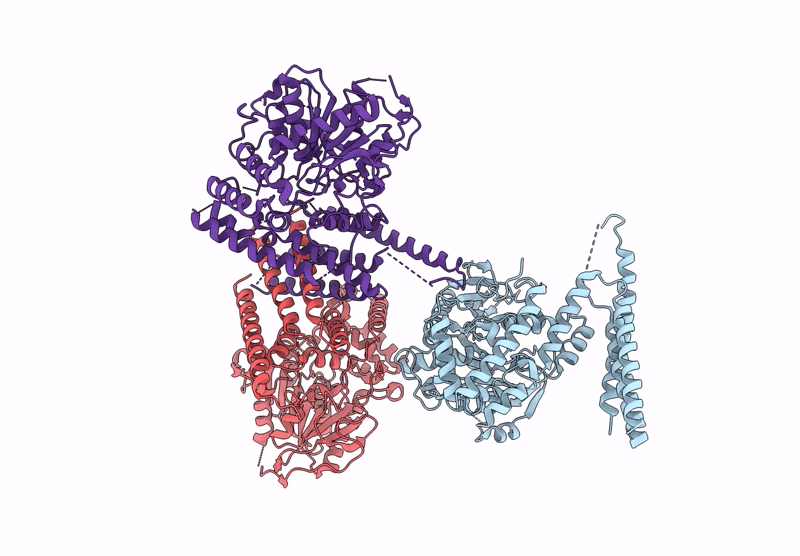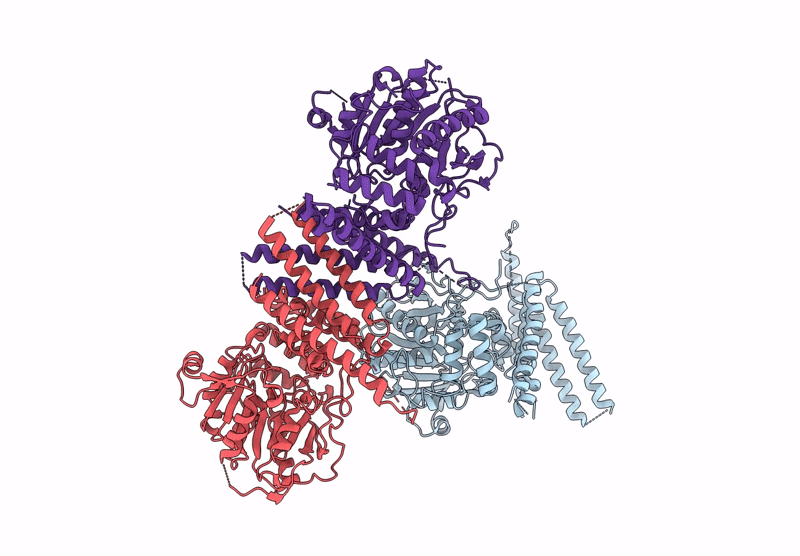
Crystal Structure Of Glycerol-Bound Full-Length Pha Synthase (Phac) From Aeromonas Caviae
Organism: Aeromonas caviae
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL |
 |
Organism: Aeromonas caviae
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of Triethylene Glycol-Bound Full-Length Pha Synthase (Phac) From Aeromonas Caviae
Organism: Aeromonas caviae
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: BIOSYNTHETIC PROTEIN Ligands: PGE |
 |
Structure Of 3-Amino-3-Carboxyltransferase In Complex With 5-Thioadenosine In The Biosynthesis Of Nocardicins
Organism: Nocardia uniformis subsp. tsuyamanensis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: A4D |
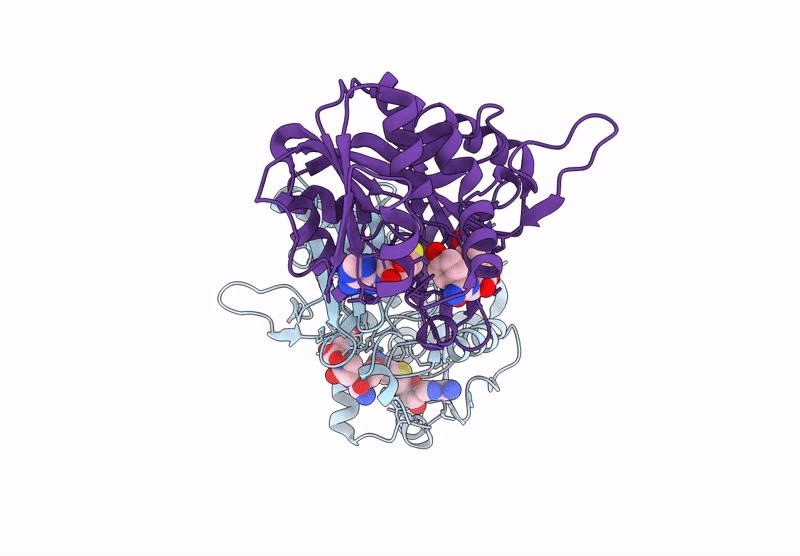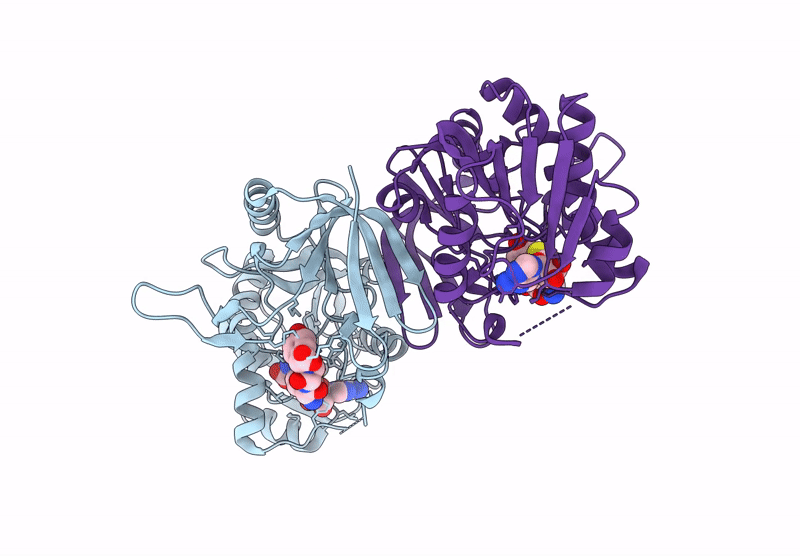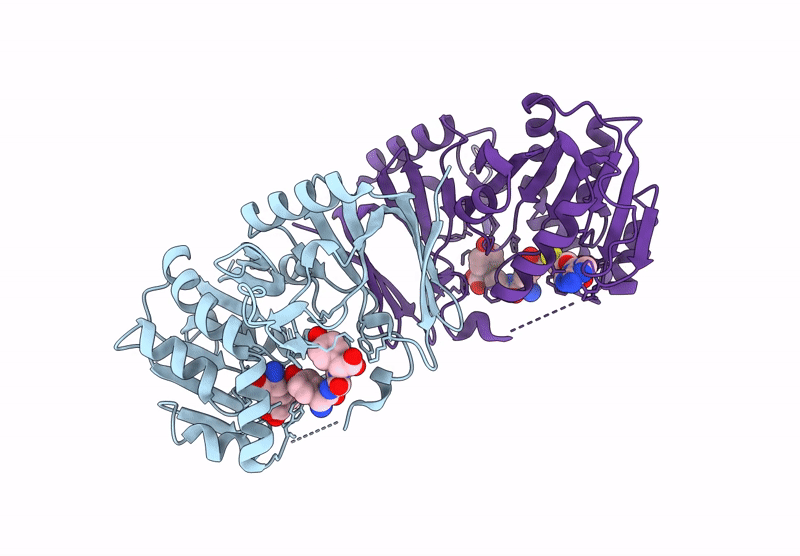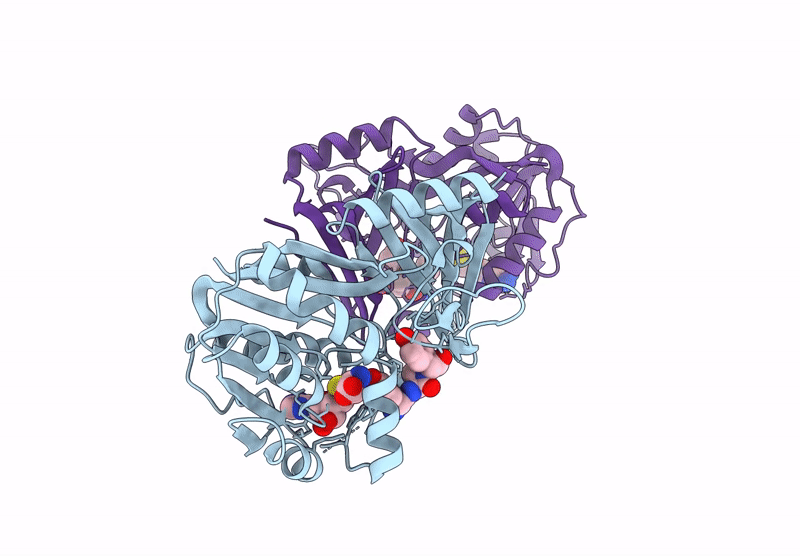 |
Structure Of 3-Amino-3-Carboxyltransferase In Complex With Sah And Nocardicin G In The Biosynthesis Of Nocardicins
Organism: Nocardia uniformis subsp. tsuyamanensis
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: SAH, A1L1U, A4D |
 |
Crystal Structure Of Nucleotide-Free Human Kinesin-1 Motor Domain (G234V Mutant)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-04-23 Classification: MOTOR PROTEIN |
 |
Crystal Structure Of Nucleotide-Free Human Kinesin-1 Motor Domain (G234A Mutant)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2025-04-23 Classification: MOTOR PROTEIN Ligands: HOH |
 |
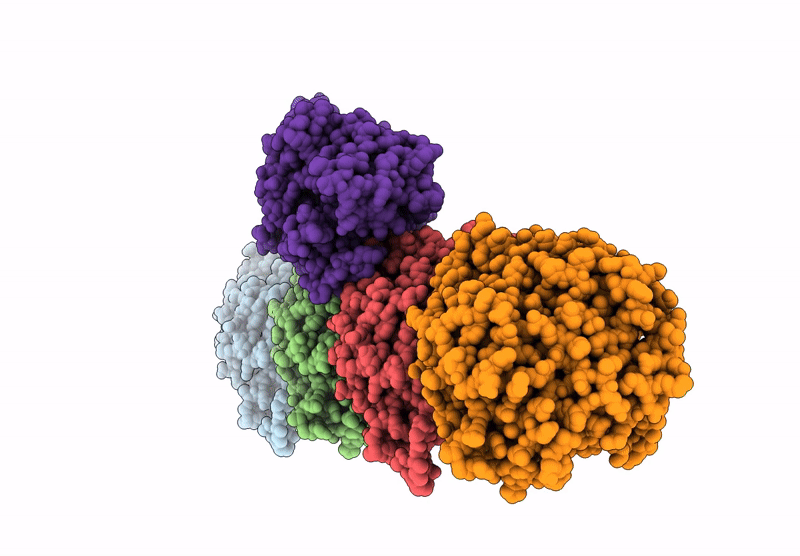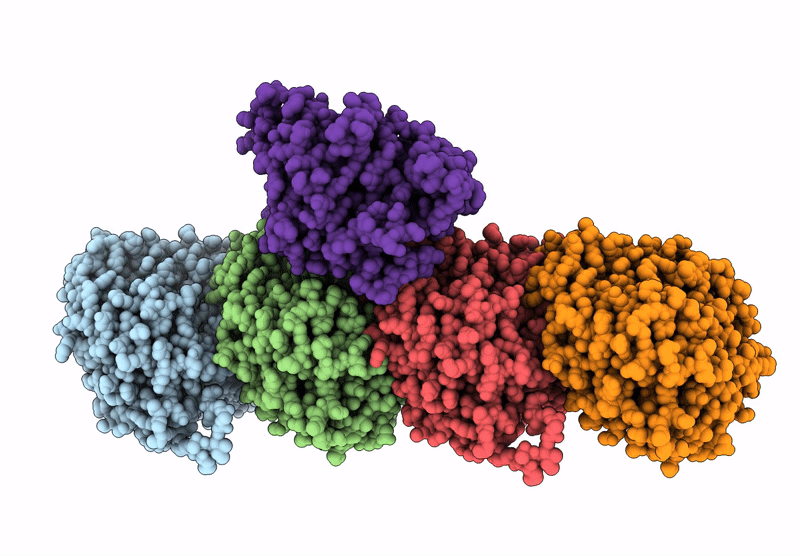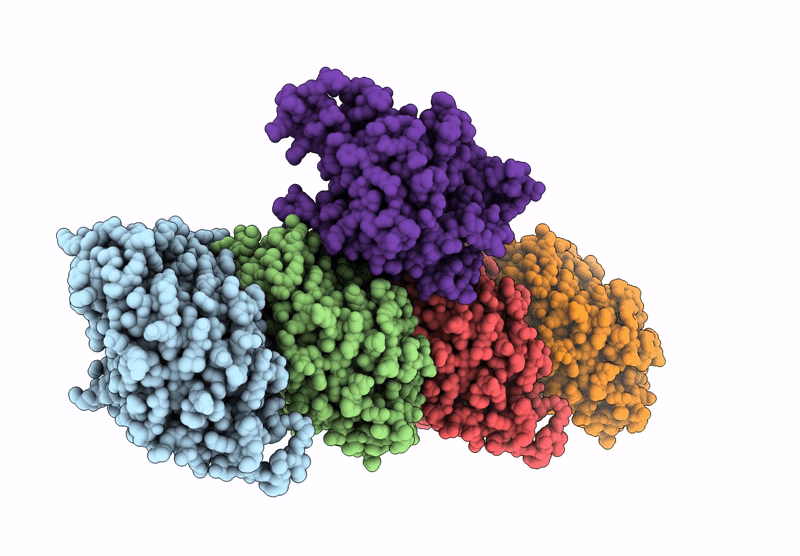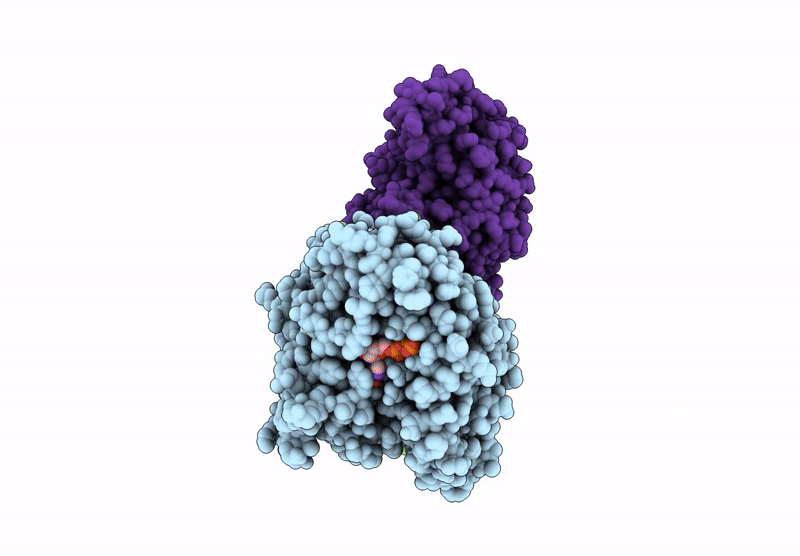
Crystal Structure Of Human Kinesin-1 Motor Domain (G234A Mutant) In Complex With Adp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-04-23 Classification: MOTOR PROTEIN Ligands: MG, ADP |
 |
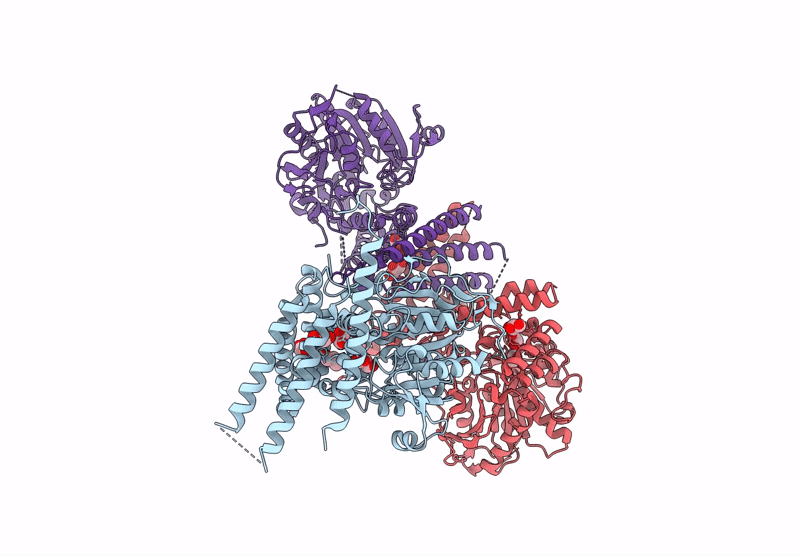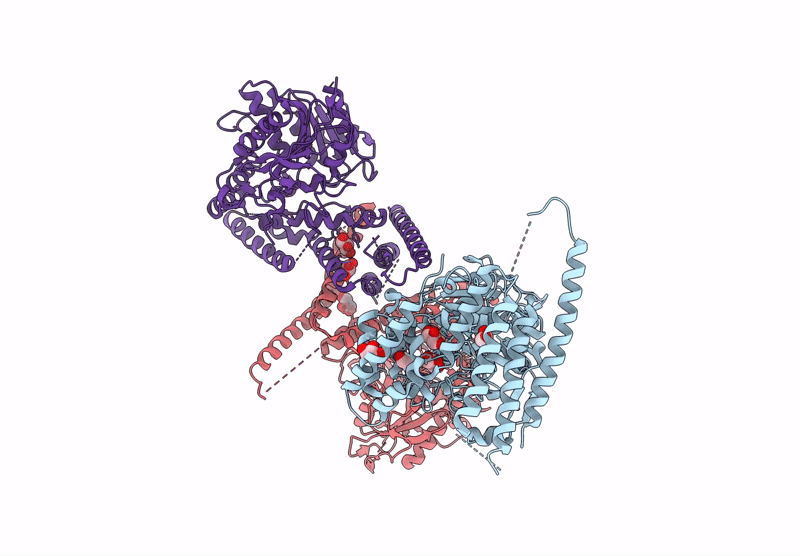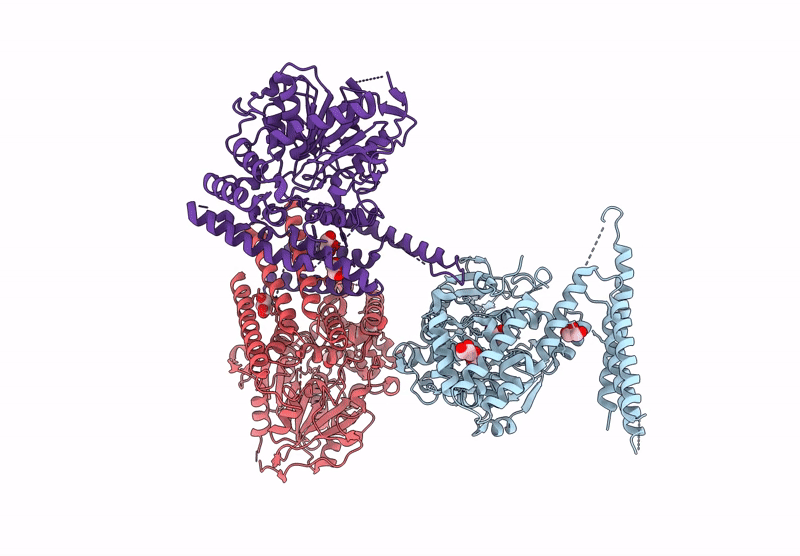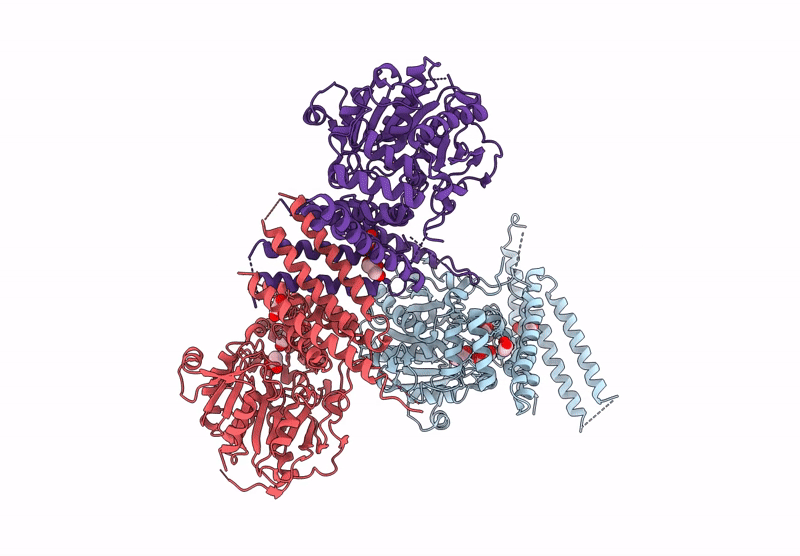
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MOTOR PROTEIN Ligands: GTP, MG |
 |
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-02-05 Classification: TRANSFERASE Ligands: CL |
 |
Complex Structure Of N-Acetyltransferase Sbzi And Carrier Protein Sbzg In The Biosynthesis Of Altemicidin
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-02-05 Classification: TRANSFERASE Ligands: SCN, CL, BCT |
 |
Organism: Streptomyces sp. atcc 700974
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-10-09 Classification: OXIDOREDUCTASE Ligands: SF4, SO4 |
 |
Crystal Structure Of Kemp Eliminase Hg3.17 In Complexed With 5-Cyanobenzotriazole
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-09-04 Classification: LYASE Ligands: VWF, SO4 |
 |
Organism: Streptomyces caelestis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-08-14 Classification: LYASE |
 |
Organism: Streptomyces lincolnensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-08-14 Classification: LYASE Ligands: GOL |
 |
Organism: Pseudomonas fluorescens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: LYASE Ligands: NAD |
 |
Organism: Pseudomonas fluorescens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: LYASE |

