Search Count: 158
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1B3B, BTB, EDO |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: A1A3N |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: A1A3O |
 |
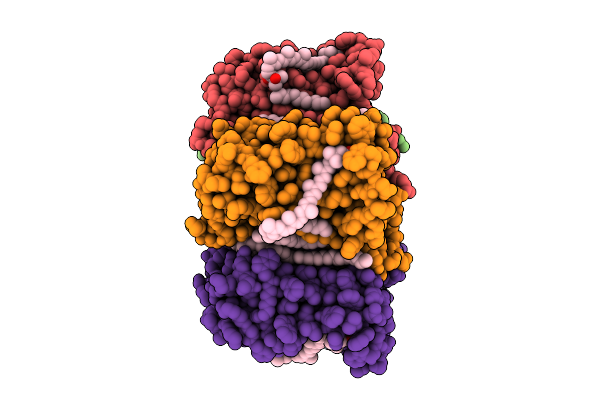
Cryo-Em Structure Of The Zeaxanthin-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, K3I |
 |
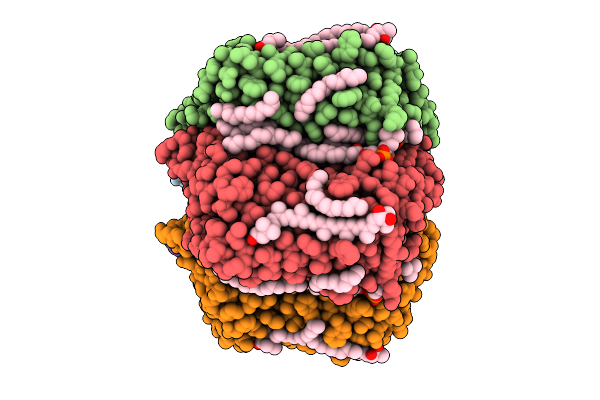
Cryo-Em Structure Of The Myxol-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, A1L4O |
 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, A1L4O, CL, PC1, PLC, R16, 8K6, D12, C14, D10 |
 |
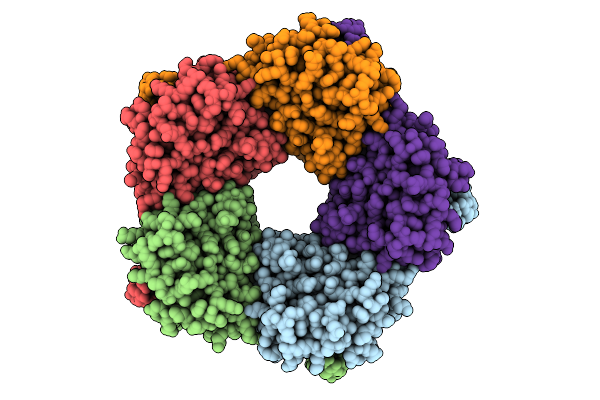
Cryo-Em Structure Of The Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, CL, PC1, PLC, D12, R16, 8K6, C14 |
 |
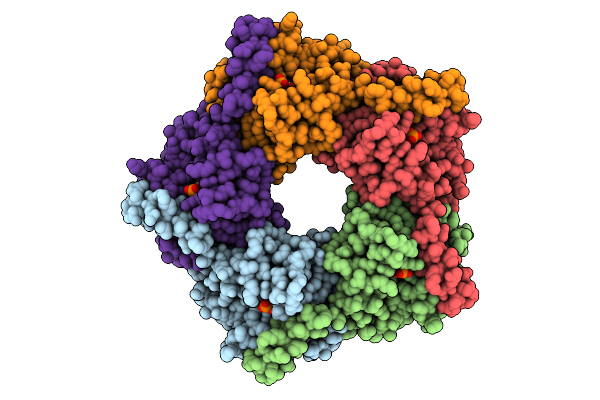
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE |
 |
X-Ray Structure Of Enterobacter Cloaca Transaldolase In Complex With D-Fructose-6-Phosphate.
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: F6R |
 |
Organism: Thermus thermophilus (strain atcc 27634 / dsm 579 / hb8)
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: SO4, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: TRANSFERASE Ligands: A1L3B, CL, GOL, EDO |
 |
Structure Of Photosynthetic Lh1-Rc Complex From The Purple Bacterium Blastochloris Tepida
Organism: Blastochloris tepida
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: PHOTOSYNTHESIS Ligands: HEC, MG, UQ8, DGA, BCB, BPB, CDL, FE, MQ7, NS5, LMT, PGV, NS0 |
 |
Organism: Pseudomonas sp. a2c
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: FMT, IMD, MG |
 |
Crystal Structure Of L-Azetidine-2-Carboxylate Hydrolase Soaked In (S)-Azetidine-2-Carboxylic Acid
Organism: Pseudomonas sp. a2c
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: 02A |
 |
Organism: Lactobacillus helveticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-03-27 Classification: HYDROLASE |
 |
Organism: Lactobacillus helveticus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-03-27 Classification: HYDROLASE |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-03-13 Classification: ISOMERASE Ligands: MN |
 |
Crystal Structure Of Lactobacillus Rhamnosus L-Rhamnose Isomerase In Complex With L-Rhamnose
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2024-03-13 Classification: ISOMERASE Ligands: MN, RM4, RAM |
 |
Crystal Structure Of Lactobacillus Rhamnosus L-Rhamnose Isomerase In Complex With D-Allulose
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2024-03-13 Classification: ISOMERASE Ligands: MN, PSJ, PSV |
 |
Crystal Structure Of Lactobacillus Rhamnosus L-Rhamnose Isomerase In Complex With D-Allose
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2024-03-13 Classification: ISOMERASE Ligands: MN, ALL, AFD |

