Search Count: 221
 |
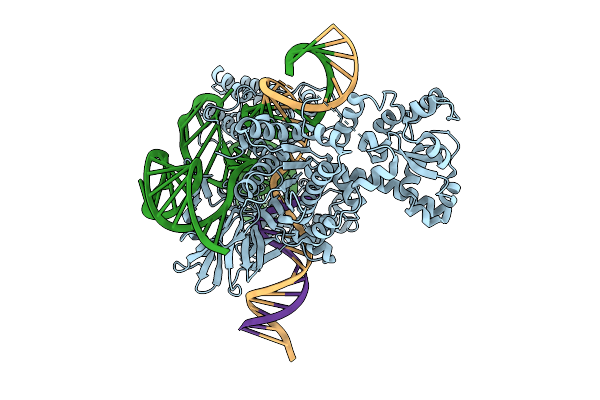
Cryo-Em Structure Of Esacas9_Nng-Guide Rna-Target Dna Complex In An Interrogation State
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN |
 |
Cryo-Em Structure Of Esacas9_Nng-Guide Rna-Target Dna Complex In An Interrogation State
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN |
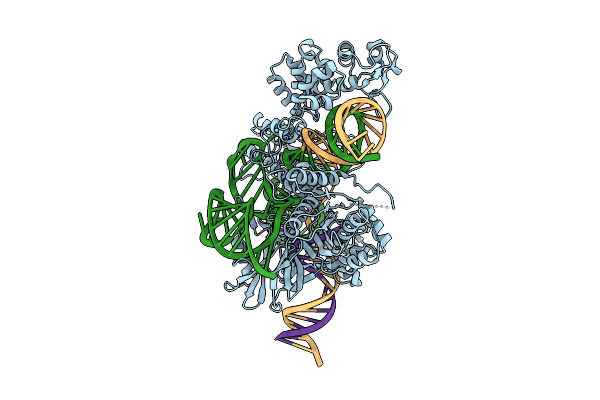 |
Cryo-Em Structure Of Esacas9_Nng-Guide Rna-Target Dna Complex In A Translocation State
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN Ligands: MG |
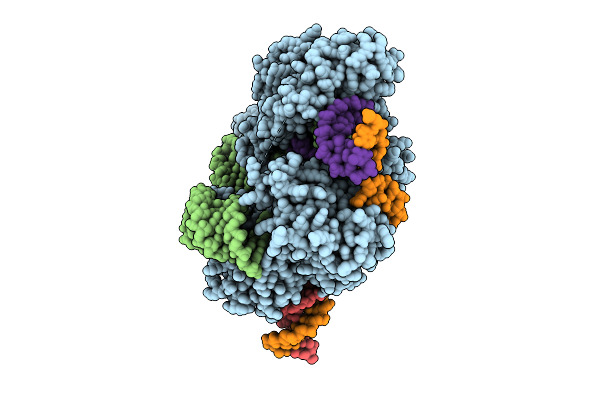 |
Cryo-Em Structure Of Esacas9_Nng-Guide Rna-Target Dna Complex In A Catalytically Active State
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN Ligands: MG |
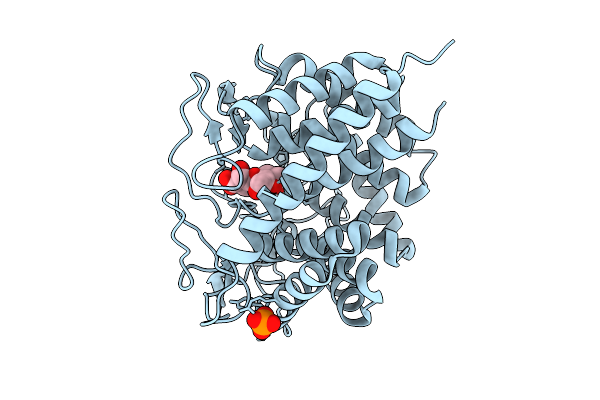
 |
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: EDO, CIT |
 |
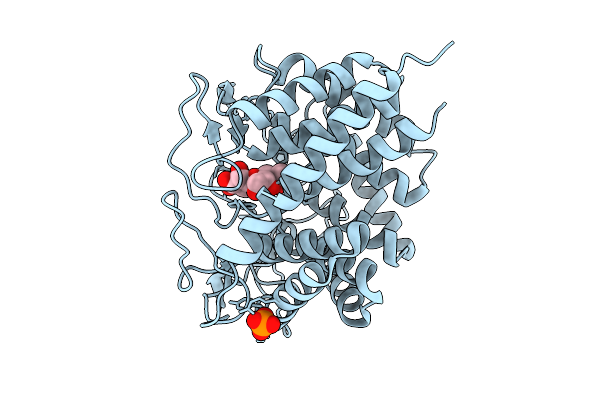
Rhodothermus Marines Cellobiose 2-Epimerase Rmce In Complex With Mannobiose
Organism: Rhodothermus marinus jcm 9785
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ISOMERASE Ligands: CL, PO4 |
 |
Rhodothermus Marines Cellobiose 2-Epimerase Rmce In Complex With Mannobiitol
Organism: Rhodothermus marinus jcm 9785
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ISOMERASE Ligands: MTL, BMA, CL, PO4 |
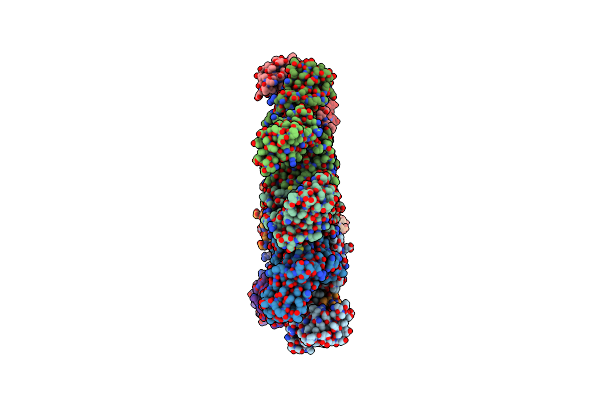 |
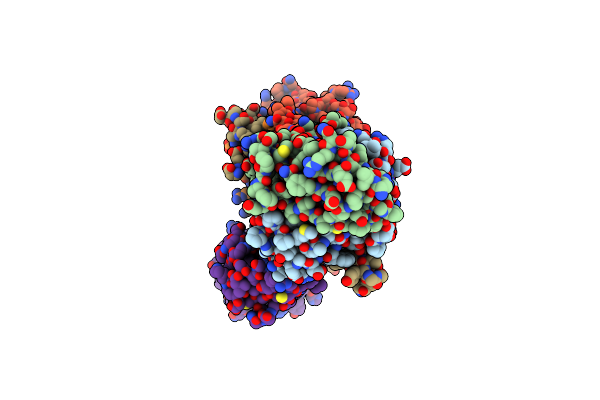
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: IMMUNE SYSTEM Ligands: CA, NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: IMMUNE SYSTEM Ligands: CA |
 |
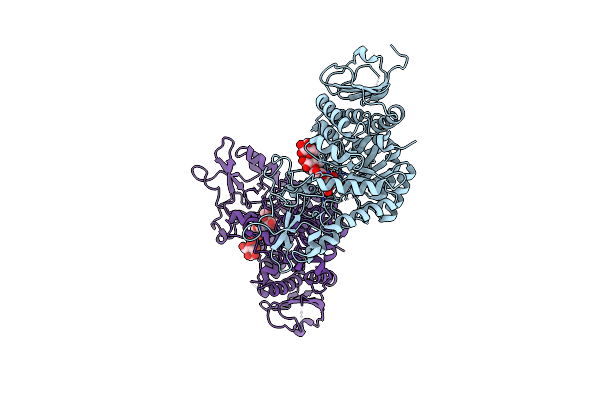
Organism: Brevibacillus laterosporus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-07-10 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: EDO, SO4, CL |
 |
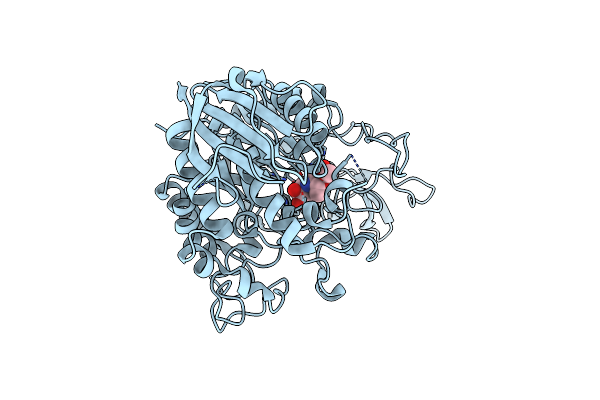
Crystal Structure Of Gh13_30 Alpha-Glucosidase Cmmb In Complex With Acarbose
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-07-03 Classification: HYDROLASE |
 |
Crystal Structure Of Gh13_30 Alpha-Glucosidase Cmmb In Complex With Acarviosin
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-03 Classification: HYDROLASE Ligands: A1L2I |
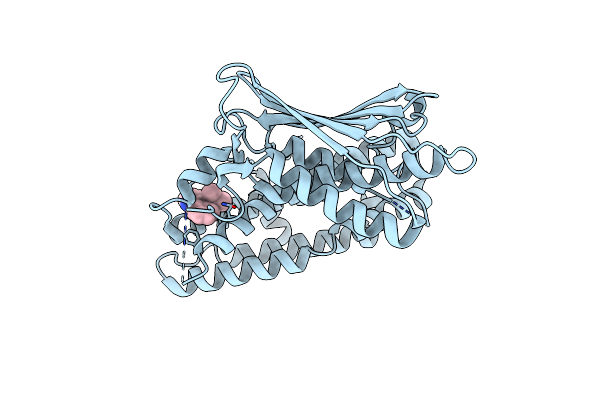 |
Structure Of Human Pyruvate Dehydrogenase Kinase 2 Complexed With Compound 6
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-06-19 Classification: TRANSFERASE Ligands: A1L16 |
 |
Structure Of Human Pyruvate Dehydrogenase Kinase 2 Complexed With Compound 16
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2024-06-19 Classification: TRANSFERASE Ligands: A1L17 |
 |
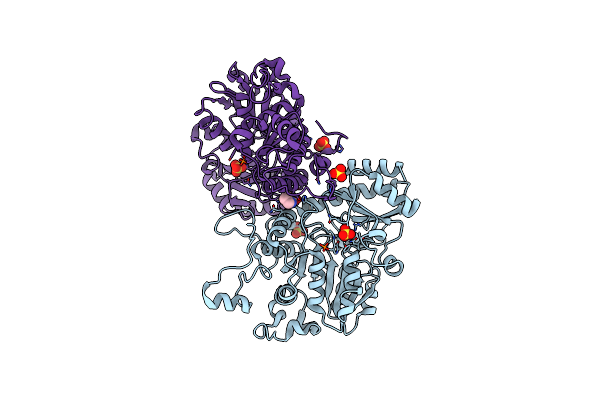
Glycoside Hydrolase Family 55 Endo-Beta-1,3-Glucanase From Microdochium Nivale
Organism: Microdochium nivale
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-04-03 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of O-Acetylhomoserine Sulfhydrylase From Lactobacillus Plantarum In The Closed Form
Organism: Lactiplantibacillus plantarum jdm1
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2024-02-14 Classification: LYASE Ligands: SO4, PRO |
 |
Crystal Structure Of O-Acetylhomoserine Sulfhydrylase From Lactobacillus Plantarum In The Open Form
Organism: Lactiplantibacillus plantarum jdm1
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-02-14 Classification: LYASE Ligands: PRO |
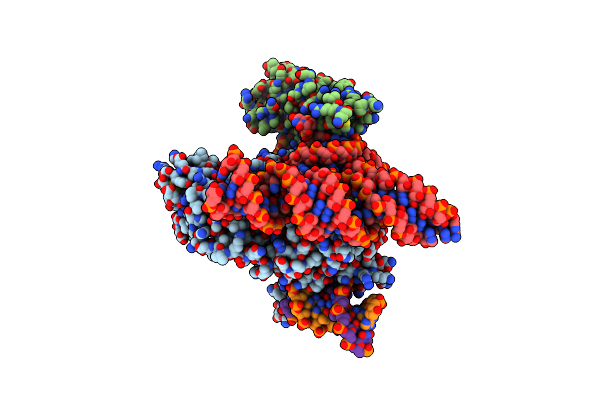 |
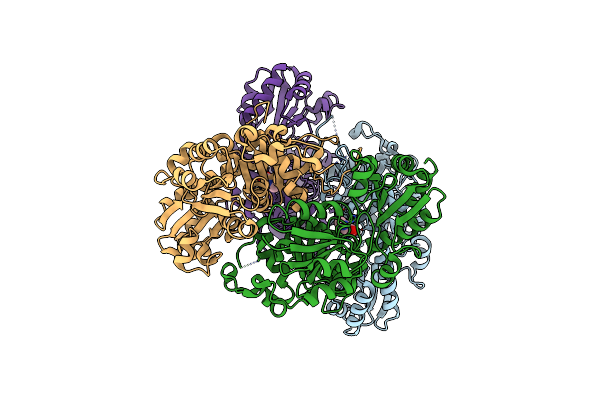
Organism: Acidibacillus sulfuroxidans, Acidibacillus sulfooxidans
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG, ZN |
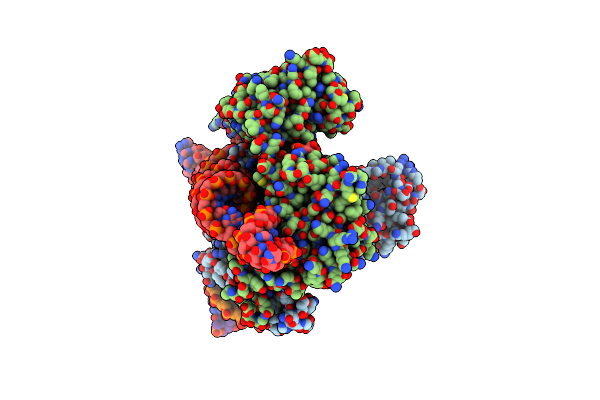 |
Organism: Sulfoacidibacillus thermotolerans
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: ZN, MG |
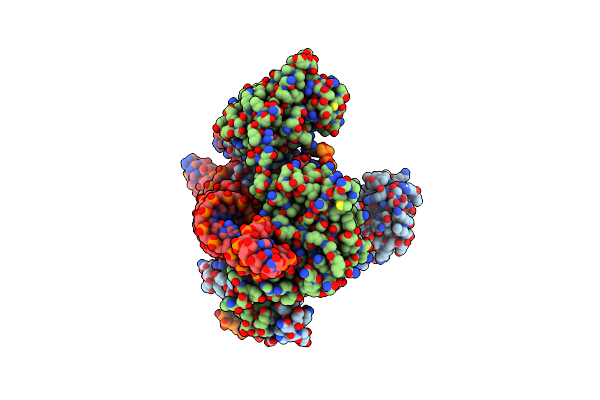 |
Organism: Sulfoacidibacillus thermotolerans
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: ZN, MG |

