Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: MEMBRANE PROTEIN Ligands: PCW, MG, A1MA7, NAG, CLR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: CELL ADHESION Ligands: CA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: CELL ADHESION |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: MEMBRANE PROTEIN Ligands: PCW, MG, PWI, NAG, CLR |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: MEMBRANE PROTEIN Ligands: PCW, MG, PXR, NAG, CLR |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: MEMBRANE PROTEIN Ligands: PCW, MG, PZ0, NAG, CLR |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: MEMBRANE PROTEIN Ligands: PCW, MG, UOU, NAG, CLR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-04-01 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: DU9, ZN, PO4, GOL |
 |
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2017-10-04 Classification: GENE REGULATION Ligands: 8TL |
 |
Structure Of Peroxiredoxin From Anaerobic Hyperthermophilic Archaeon Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2013-07-10 Classification: OXIDOREDUCTASE Ligands: FLC |
 |
Archaeal Non-Discriminating Glutamyl-Trna Synthetase From Methanothermobacter Thermautotrophicus
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2010-08-04 Classification: LIGASE Ligands: ACY, EDO, ZN, CA |
 |
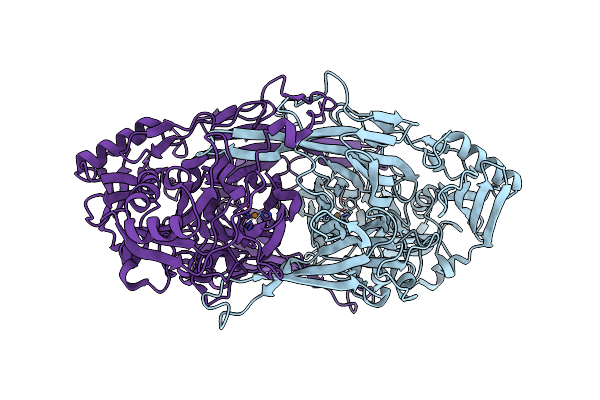
Crystal Structure Of Copper Amine Oxidase From Arthrobacter Globiformis: Initial Intermediate In Topaquinone Biogenesis
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2002-08-07 Classification: OXIDOREDUCTASE Ligands: CU |
 |
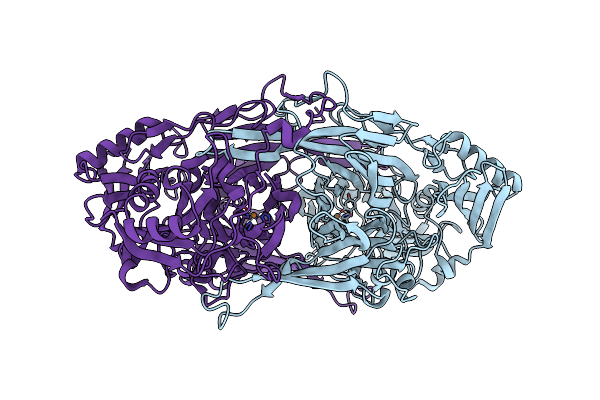
Crystal Structure Of Copper Amine Oxidase From Arthrobacter Globiformis: Early Intermediate In Topaquinone Biogenesis
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Release Date: 2002-08-07 Classification: OXIDOREDUCTASE Ligands: CU |
 |
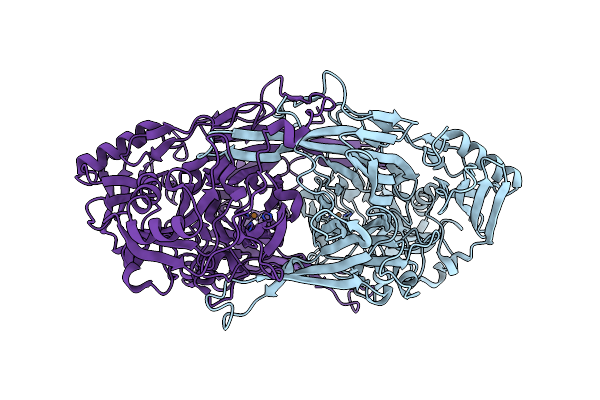
Crystal Structure Of Copper Amine Oxidase From Arthrobacter Globiformis: Late Intermediate In Topaquinone Biogenesis
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2002-08-07 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Crystal Structure Of Copper Amine Oxidase From Arthrobacter Globiformis: Holo Form Generated By Biogenesis In Crystal.
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2002-08-07 Classification: OXIDOREDUCTASE Ligands: CU |