Search Count: 23
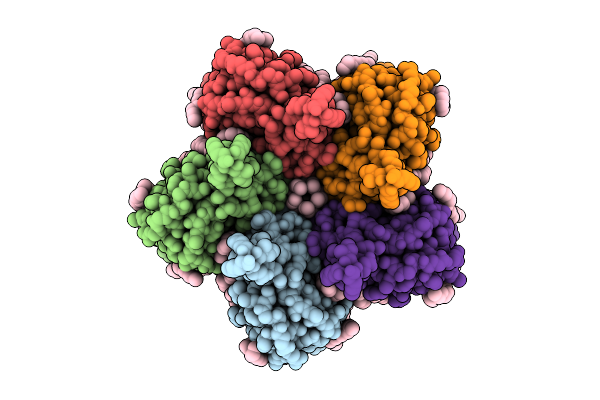 |
Cryo-Em Structure Of The Flotillin-Associated Rhodopsin Psfar In Detergent Micelle
Organism: Candidatus pseudothioglobus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA |
 |
Cryo-Em Structure Of The Light-Driven Proton Pump Pspr In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET, LMT |
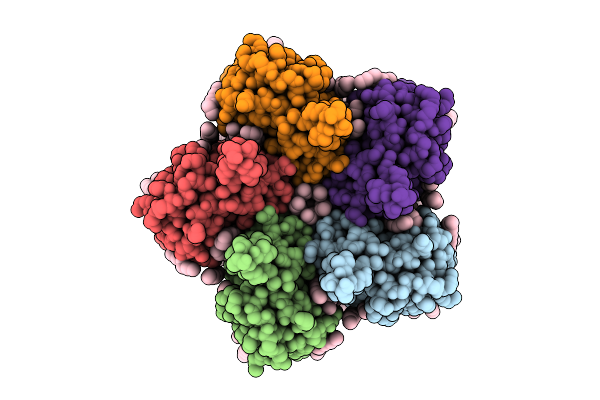 |
Cryo-Em Structure Of The Double Mutant H84V/E120G Of The Flotillin-Associated Rhodopsin Psfar In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
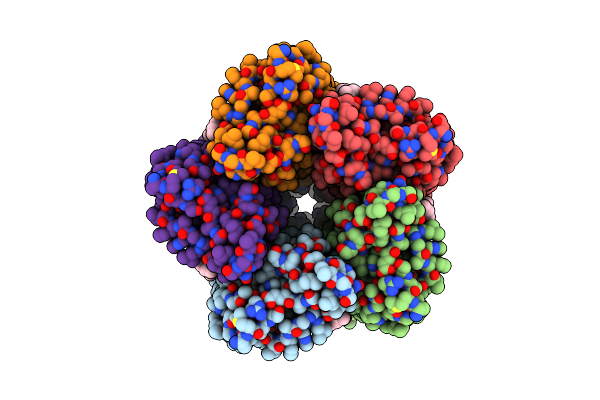
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
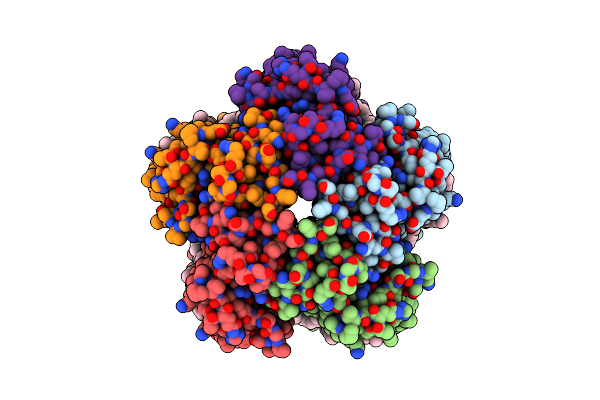
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LMT, LFA, RET |
 |
Cryo-Em Structure Of The Microbial Rhodopsin Cryor1 At Ph 10.5 In Detergent In The Ground State
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LMT, RET, LFA |
 |
Cryo-Em Structure Of The Microbial Rhodopsin Cryor1 At Ph 10.5 In Detergent In The M State
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LMT, RET |
 |
Organism: Subtercola endophyticus
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: HYDROLASE Ligands: A1L1M, PEG, GOL |
 |
Crystal Structure Of Rat Glutathione Transferase Omega 1 Bound To Glutathione
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-07-31 Classification: TRANSFERASE Ligands: GSH |
 |
Organism: Arabidopsis thaliana, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-06-28 Classification: SIGNALING PROTEIN |
 |
 |
Organism: Actinobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-12-11 Classification: MEMBRANE PROTEIN Ligands: LFA, OLC, SO4, RET, GOL, OLA |
 |
Organism: Actinobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-12-11 Classification: MEMBRANE PROTEIN Ligands: ACT, OLA, GOL, OLC, LFA, SO4, RET |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2019-08-28 Classification: ANTIMICROBIAL PROTEIN |
 |
Structure Of A New Shkt Peptide From The Sea Anemone Oulactis Sp: Osptx2A-P2
|
 |
Structure Of A New Shkt Peptide From The Sea Anemone Oulactis Sp: Osptx2A-P1
|
 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2009-03-03 Classification: TRANSFERASE |

