Search Count: 18
 |
Pseudomonas Fluorescens G150T Isocyanide Hydratase At 298 K Xfel Data, Free Enzyme
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2024-02-21 Classification: LYASE Ligands: CL |
 |
Pseudomonas Fluorescens G150T Isocyanide Hydratase At 298 K Xfel Data, Thioimidate Intermediate
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2024-02-21 Classification: LYASE Ligands: QCV, CL |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2023-09-20 Classification: LYASE Ligands: CL |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2023-09-20 Classification: LYASE Ligands: CL |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2023-09-20 Classification: LYASE Ligands: CL |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-09-20 Classification: LYASE Ligands: CL |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-09-20 Classification: LYASE |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-09-20 Classification: LYASE |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2023-09-20 Classification: LYASE Ligands: EDO |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2023-09-20 Classification: LYASE Ligands: EDO |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2023-09-20 Classification: LYASE Ligands: EDO |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-10-13 Classification: LYASE Ligands: MN, GTP, CL |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2021-10-13 Classification: LYASE Ligands: MN, NA, PEP, GDP, CO2 |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2021-10-13 Classification: LYASE Ligands: CO2, GDP, PEP, MN |
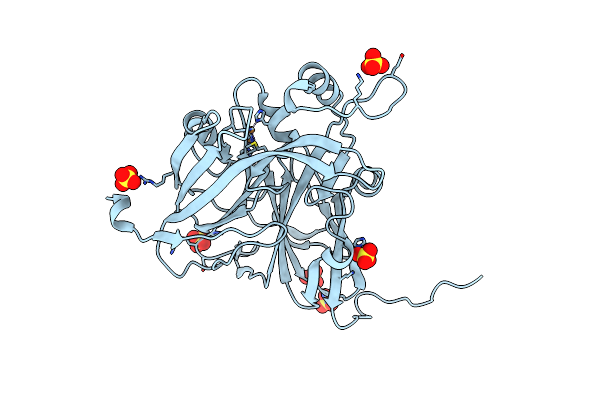 |
Serial Cu Nitrite Reductase Structures At Elevated Cryogenic Temperature, 240K. Dataset 1.
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2017-07-26 Classification: OXIDOREDUCTASE Ligands: CU, SO4 |
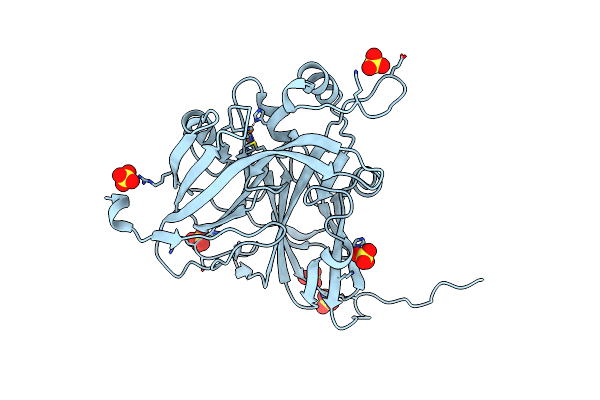 |
Serial Cu Nitrite Reductase Structures At Elevated Cryogenic Temperature, 240K. Dataset 2.
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2017-07-26 Classification: OXIDOREDUCTASE Ligands: CU, SO4 |
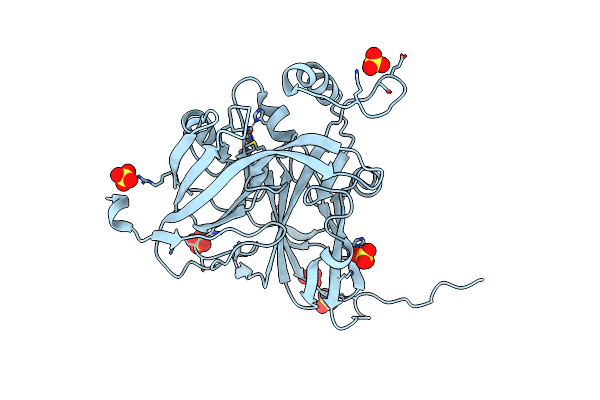 |
Serial Cu Nitrite Reductase Structures At Elevated Cryogenic Temperature, 240K. Dataset 3.
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-07-26 Classification: OXIDOREDUCTASE Ligands: CU, SO4 |
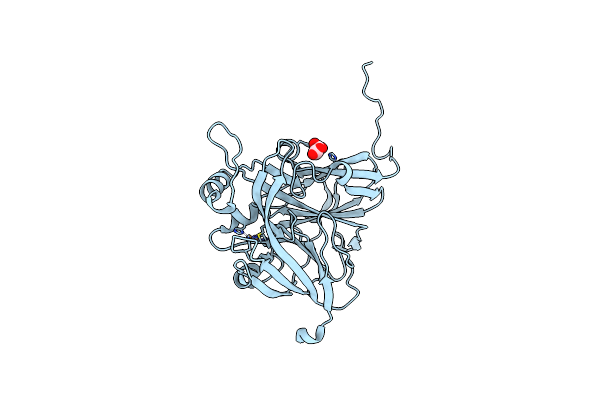 |
Serial Cu Nitrite Reductase Structures At Elevated Cryogenic Temperature, 100K Reference Dataset.
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-07-26 Classification: OXIDOREDUCTASE Ligands: CU, MLI |

